Research
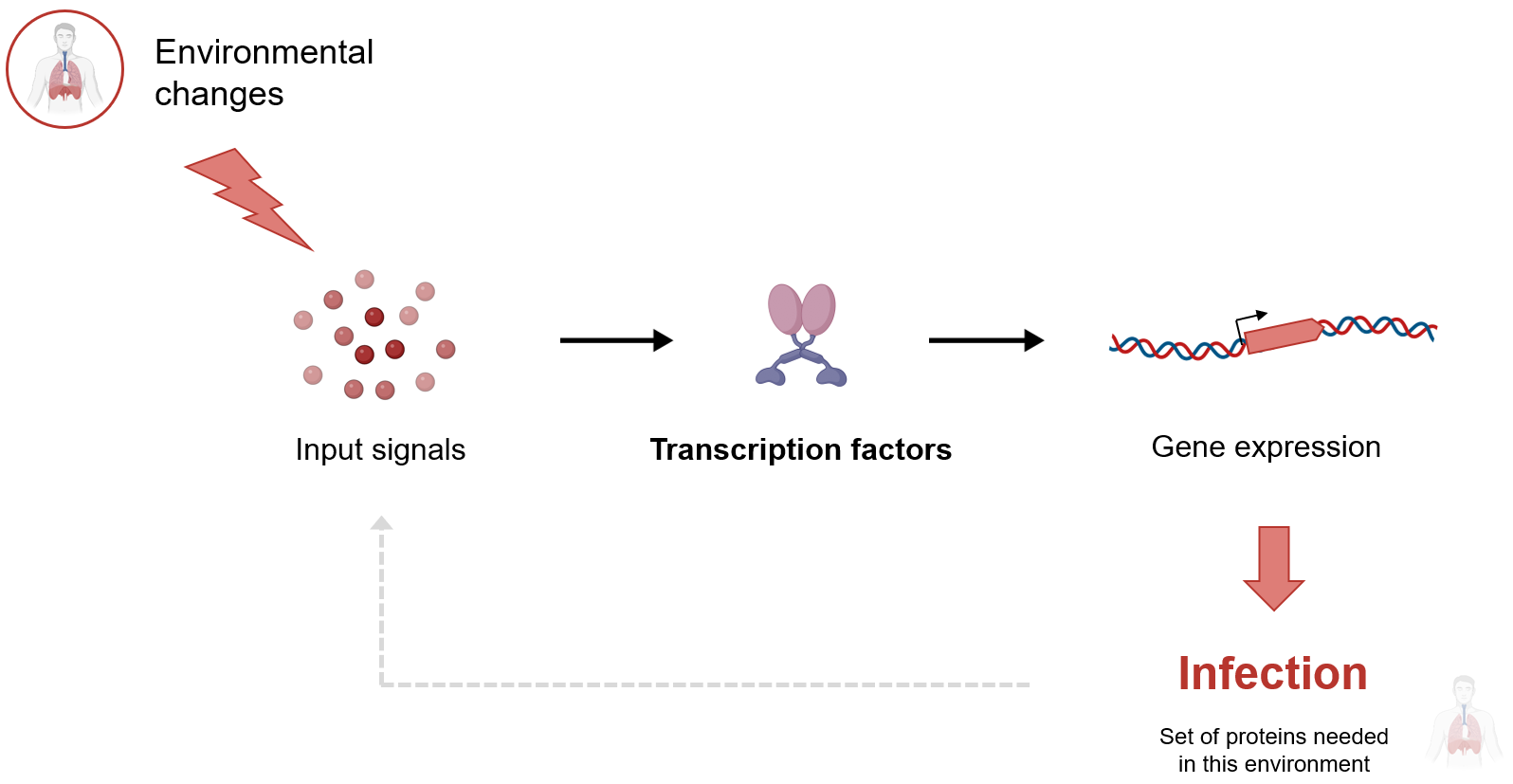
Complex networks of transcription factors allow cells to adapt to changes in their environement through (i) the sensing of external signals, and (ii) corresponding gene expression responses. Despite being critical for physiological adaptation, these networks are still poorly characterized, making it virtually impossible to accurately predict gene expression responses or identify key regulatory processes driving adaptation to environmental changes.
The research in our group aims at getting a mechanistic understanding of how bacteria use transcriptional regulation to adapt to environmental changes. We work at the scales of molecular interactions and networks, focusing on transcription factors - the DNA-binding proteins that regulate gene expression. To that aim, we use and develop high-throughput sequencing-based approaches to characterize transcriptional regulatory networks. These include automated pipelines for characterization of transcription factor interactions at the scale of entire regulatory networks.
Research topics:
1. Regulatory networks in bacterial pathogens
Here, we focus on bacterial pathogens to understand how transcriptional regulatory networks evolve to integrate new genetic material, and how they enable the physiological adaptation needed for infections. To address this question, we leverage our unique high-throughput capabilities to map regulatory networks at scale. Using these networks, we can interpret data from infections to identify and characterize molecular interactions that allow bacterial pathogens to adapt to the human body and cause infections. Additionally, we study regulatory network evolution within and between pathogenic species to identify core regulatory interactions conserved across pathogenic isolates and reveal how changes in gene content reshape regulatory networks. Currently, our research is focused on two key bacterial pathogens: Escherichia coli and Pseudomonas aeruginosa.
2. Molecular rules of transcription factor interactions
Here, we aim to define transferable molecular rules that dictate transcription factor regulatory interactions to enable the prediction and manipulation of gene expression responses. To that aim, we combine high-throughput protein-DNA binding assays with computational modeling to decode and predict transcription factors DNA-binding specificity. Using these tools, we aim to investigate how DNA-binding preferences evolve between species to bridge the gap between TF sequence diversity and function. Additionally, we aim to explore the use of such predictive tools for the design of new-to-nature transcription factors with on-demand functions.
References
- Trouillon, J., Huber, A. E., Trabesinger, Y., & Sauer, U. (2025). Predicting input signals of transcription factors in Escherichia coli. Molecular Systems Biology, 1-17.
- Trouillon, J., Doubleday, P. F., & Sauer, U. (2023). Genomic footprinting uncovers global transcription factor responses to amino acids in Escherichia coli. Cell Systems, 14(10), 860-871.
- Holbrook-Smith, D., Trouillon, J., & Sauer, U. (2023). Metabolomics and Microbial Metabolism: Toward a Systematic Understanding. Annual Review of Biophysics, 53.