anNET
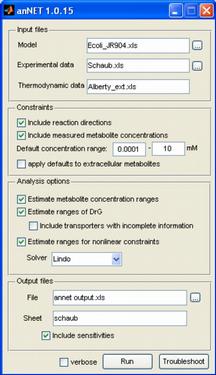
anNET affords a generalized framework for Network Embedded Thermodynamic (NET) analysis of quantitative metabolomic datasets. Building upon principles of thermodynamics, this method allows for a quality check of measured metabolite concentrations and enables to spot metabolic reactions where active regulation potentially controls metabolic flux.
The applications of anNET are multiple, for example:
- Check thermodynamic consistency
- Estimate unmeasured concentrations
- Resolve concentrations in different compartments
- Find minimum/maximum feasible NADH/NAD ratio or adenylate energy charge
- Infer reaction direction
- Verify reversibility in model
- Spot putative control sites
- Exclude activity of transporters
Requirements
Matlab 6.0 or higher (tested on Windows and Linux)
Matlab Optimization Toolbox 3 (The Mathworks) or Lindo API (Lindo Systems, Inc.)
References
- Kümmel, Panke, and Heinemann, Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol Syst Biol. 2006;2:2006.0034. external page Pubmed
- Zamboni, Kümmel and Heinemann, anNET: a tool for network-embedded thermodynamic analysis of quantitative metabolome data, BMC Bioinformatics 2008, 9:199. external page BMC