2015
Two new papers published by our collaborators
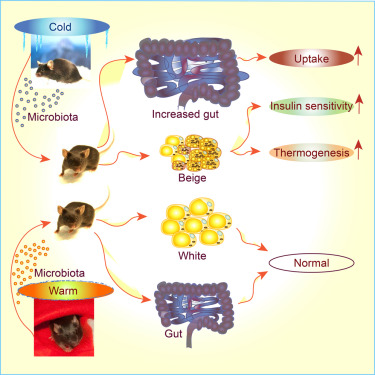
We congratulate with Prof. Trajkovski (U. Geneva) for demonstrating how "Gut Microbiota Orchestrates Energy Homeostasis during Cold" and Prof Ristow (ETH Zurich) to demonstrate the role of branched chain amino acids in regulating aging.
Ulrich Genick awarded with “Golden Owl” of the VSETH

for outstanding contributions to teaching. Congratulations!
Luca Del Medico starts his PhD
Luca performed his master thesis in our group and now joined the Christen lab for his PhD. Luca will work on novel systems and synthetic biology approaches to study core networks in prokaryotic organisms.
Pseudo-transition Analysis Identifies the Key Regulators of Dynamic Metabolic Adaptations from Steady-State Data
The Sauer Lab developed an experimental-computational approach to identify regulatory needles in metabolic haystacks. By using steady-state 13C flux, metabolites and transcript data, the approach revealed the sparse and transition-dependent regulators that drive transitions between carbon sources in Escherichia coli. Thus, pseudo-transition analysis is a novel approach to efficiently explore the vast regulatory landscape of dynamic adaptations using relatively few stationary observations.
Wollscheid Lab: Master Thesis in Receptor Nanoscale Organisation & Cellular Signaling
The research of our lab focuses on cell surface protein in general and specifically on receptors. We recently suceeded in developing a technology which enables the decoding of ligand-receptor interactions [1].
Christen Lab: New openings for Master students
Mapping cellular core functions by transposon mutagenesis and next generation sequencing
Paola named EMBO Young Investigator

Paola Picotti was selected as EMBO Young Investigator. Congratulations!
US patent issued!
Our US patent 9150916 on “Compositions and methods for identifying the essential genome of an organism” was issued on Oct. 6th 2015. The Method relates to our PCR based transposon sequencing (TnSeq) strategy to define essential DNA parts of an organism with base pair resolution.
We welcome Martin Spillmann and Donat Appert
Martin joined the Christen group for his master thesis and Donat started his semester project in our lab. Welcome on board!
Dynamic exometabolome analysis reveals active metabolic pathways in non-replicating mycobacteria
In a collaborative effort of the Sauer and Zamboni labs at IMSB and the Berry Lab at NIH, Michael and his co-workers integrated dynamic changes in the secretion and uptake of metabolites with genome-scale metabolic network reconstructions and identified active metabolic routes necessary for mycobacteria survival under hypoxic conditions.
Welcome to Valentina Cappelletti
Welcome to Valentina Cappelletti who will visit our lab for several months.
Targeted Proteomics course registration is open!
In this course we offer theoretical introductory lectures, comprehensive hands-on training and discussion rounds on each of the typical tasks performed in targeted proteomics workflows. In previous years the focus of the course has been entirely on SRM (selected reaction monitoring). This year the course content is being expanded to include other methods such as PRM (parallel reaction monitoring) and DIA (data independent acquisition), with a particular focus on SWATH MS.
Aebersold lab: Master thesis/semester project in Structural Proteomics
We are looking for motivated semester/master students to join our structural proteomics team at ETH to work at the interface between proteomics and structural biology.
Sauer Group, Metabolism, Systems Biology, Modelling, Bistability
Understanding history-dependence (hysteresis) of growth states in E. coli
Real-time metabolome profiling of the metabolic switch between starvation and growth
The Zamboni and Sauer Lab developed a platform for real-time metabolome profiling by directly injecting living bacteria, yeast or higher cell suspensions into a high-resolution time-of-flight mass spectrometer. We congratulate Hannes and all the coauthors.
Posttranslational regulation of microbial metabolism
In a review published in "Current opinion in Microbiology“, we discuss the division-of-labor between the different regulatory layers in making microbial metabolic decisions, and highlight the current challenges we are facing when elucidating the role of posttranslational regulation.
The outer mucus layer hosts a distinct intestinal microbial niche
In collaboration with the Sauer lab, Hai Li & Julien P. Limenitakis from the Maurice Müller Laboratories (DKF), University of Bern, describe in detail the compositional differences between lumen and outer mucus layer of the intestine in mice and its consequence for the present microbiota.
Cyclic operation of the pentose phosphate, Entner-Doudoroff and glycolytic pathways in Pseudomonas putida
In collaboration with the Sauer lab, Pablo Ivan Nikel from the Centro Nacional de Biotecnologia (CNB-CSIC, Spain), describes a cyclic operation of the pentose phosphate, Entner-Doudoroff and glycolytic pathways in Pseudomonas putida.
Aebersold lab, student (master) project in phosphopeptide enrichment.
We are looking for students who would like to join project for development of the new phosphopeptide enrichent for the analysis of small sample amounts. These projects are the perfect opportunity to get hands-on experience with proteomics methods and mass spectrometry instrumentation. Prior experience with mass spectrometry is not essential, although candidates with basic knowledge of proteomics methods are preferred.
We welcome Amelia Palermo!
Amelia joined the lab as an Academic Guest from the Università La Sapienza of Rome to exploit non-targeted metabolomics in the context of doping.