Publications List
Selected Publications
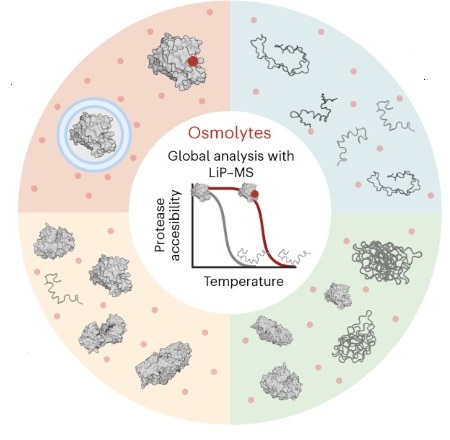
M. Pepelnjak, B. Velten, N. Näpflin, T. von Rosen, U.C. Palmiero, J.H. Ko, H.D. Maynard, P. Arosio, E. Weber-Ban, N. de Souza, W. Huber, P. Picotti. external page In situ analysis of osmolyte mechanisms of proteome thermal stabilization. (2024) Nat. Chem. Biol., Feb 29. 2024

L. Malinovska, V. Cappelletti, D. Kohler, I. Piazza, T.H. Tsai, M. Pepelnjak, P. Stalder, C. Dörig, F. Sesterhenn, F. Elsässer, Kralickova L., Beaton N., Reiter L., N. de Souza, O. Vitek, P. Picotti. external page Proteome-wide structural changes measured with limited proteolysis-mass spectrometry: an advanced protocol for high-throughput applications. (2023) Nat Protoc., 18(3):659-682.

M.T. Mackmull, L. Nagel, F. Sesterhenn, J. Muntel, J. Grossbach, P. Stalder, R. Bruderer, L. Reiter, W.D.J. van de Berg, N. de Souza, A. Beyer, P. Picotti. external page Global, in situ analysis of the structural proteome in individuals with Parkinson's disease to identify a new class of biomarker. (2022) Nat. Struct. Mol. Biol., 29(10):978-989.
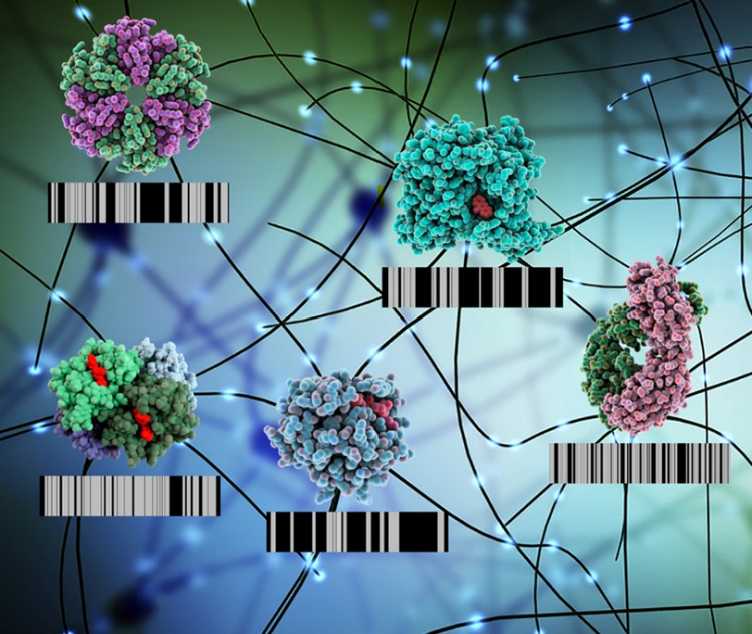
V. Cappelletti, T. Hauser, I. Piazza, M. Pepelnjak, L. Malinovska, T. Fuhrer, Y. Li, C. Dörig, P. Boersema, L. Gillet, J. Grossbach, A. Dugourd, J. Saez-Rodriguez, A. Beyer, N. Zamboni, A. Caflisch, N. de Souza, P. Picotti. external page Dynamic 3D proteomes reveal protein functional alterations at high resolution in situ. (2021) Cell, 184 (2), 545-559. e22.

I. Piazza, N. Beaton, R. Bruderer, T. Knobloch, C. Barbisan, L. Chandat, A. Sudau, I. Siepe, O. Rinner, N. de Souza, P. Picotti, L. Reiter. external page A machine learning-based chemoproteomic approach to identify drug targets and binding sites in complex proteomes. (2020) Nat. Commun., 11 (1), 1-13.

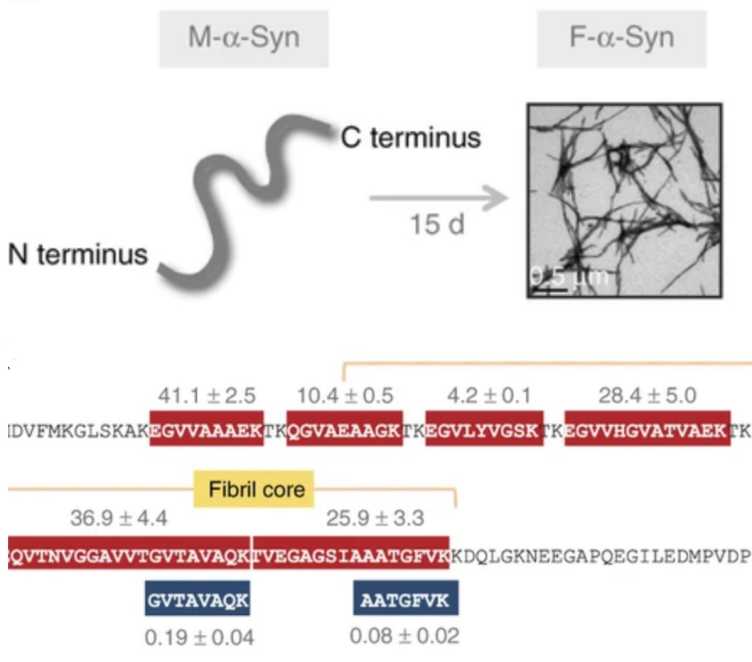
M. Soste, K. Charmpi, F. Lampert, J.A. Gerez, M. van Oostrum, L. Malinovska, P.J. Boersema, N.C. Prymaczok, R. Riek, M. Peter, S. Vanni, A. Beyer, P. Picotti. external page Proteomics-Based Monitoring of Pathway Activity Reveals that Blocking Diacylglycerol Biosynthesis Rescues from Alpha-Synuclein Toxiciexternal page ty. Cell Syst. 2019 Sep 25;9(3):309-320.e8.
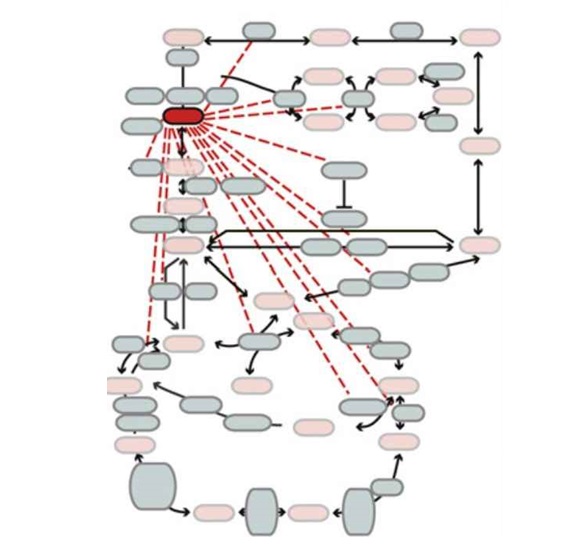
I. Piazza, K. Kochanowski, V. Cappelletti, T. Fuhrer, E. Noor, U. Sauer, P. Picotti. external page A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication. (2018) Cell, 172 (1-2), 358-372. e23.

S. Schopper, A. Kahraman, P. Leuenberger, Y. Feng, I. Piazza, O. Müller, P.J. Boersema, P. Picotti. external page Measuring protein structural changes on a proteome-wide scale using limited proteolysis-coupled mass spectrometry. (2017) Nat. Protoc.., 12 (11), 2391-2410.
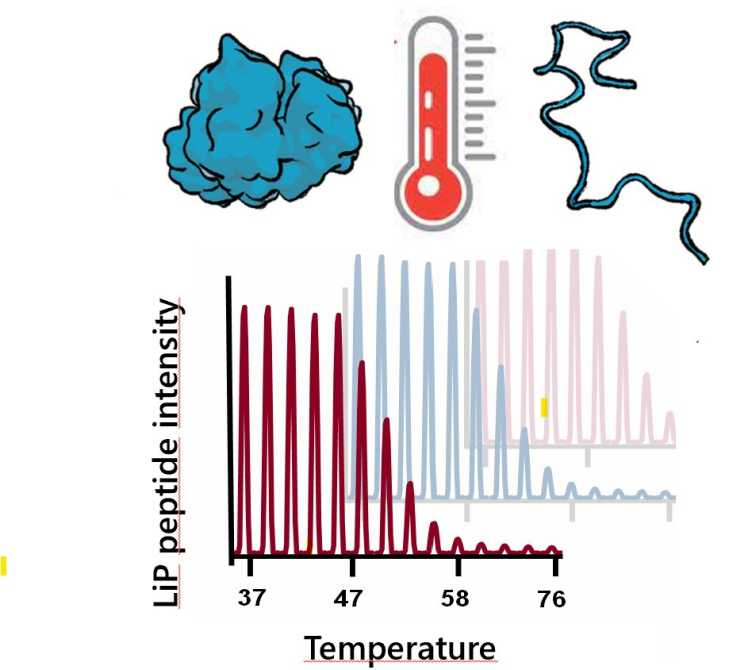
P. Leuenberger, S. Ganscha, A. Kahraman, V. Cappelletti, P.J. Boersema, C. von Mering, M. Claassen, P. Picotti. external page Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. (2017) Science, 355 (6327), eaai7825.
Y. Feng, G. De Franceschi, A. Kahraman, M. Soste, A. Melnik, P.J. Boersema, P.P. de Laureto, Y. Nikolaev, A.P. Oliveira, P. Picotti. external page Global analysis of protein structural changes in complex proteomes. (2014) Nat. Biotechnol., 32 (10), 1036-1044.

M. Soste, R. Hrabakova, S. Wanka, A. Melnik, P. Boersema, A. Maiolica, T. Wernas, M. Tognetti, C. von Mering, P. Picotti. external page A sentinel protein assay for simultaneously quantifying cellular processes. Nat Methods. 2014 Oct;11(10):1045-8.