Claassen group, Computational Systems Biology
Ab initio cell type definition from high-dimensional single-cell data
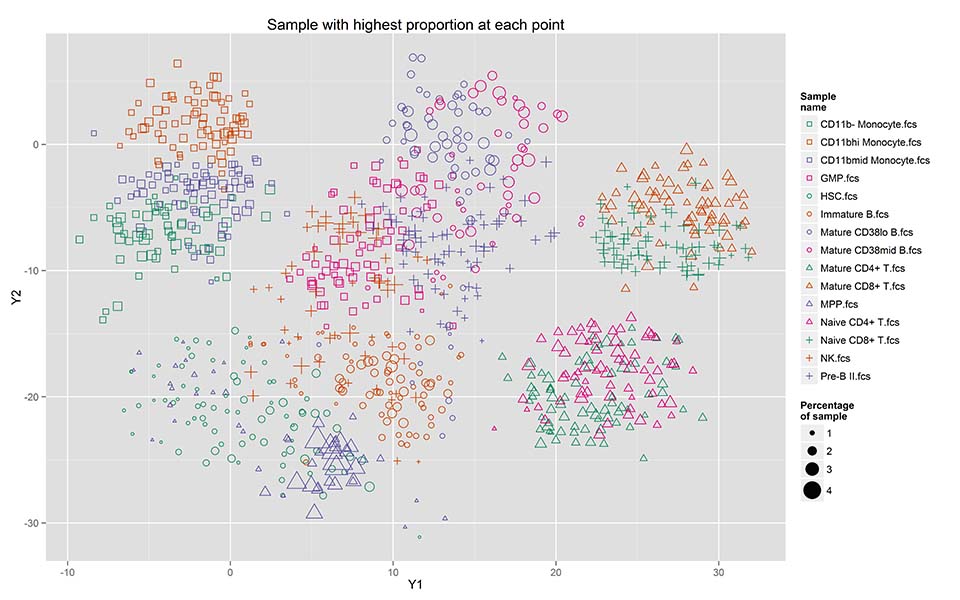
Recent technical advances in single cell proteomics allow for simultaneous quantification of up to 50 proteins levels [1], provide rich data sources to characterize canonical as well as novel cell types in health and disease states such in immune and cancer biology. Definition of cell subtypes in single-cell data has typically been performed by supervised techniques that are based on carefully selected cell surface markers. Recent unsupervised learning techniques aim at defining novel cell type defining markers in a data driven fashion [2,3]. However, these approaches have limitations such as lack of robustness in outputs, and potentially poor separation of subtypes. We address these issues by developing approaches that incorporate prior knowledge about biochemical and differentiation processes that give rise to the observed cell type diversity. Possible semester projects in this area include:
- Assessing different formal approaches to account for prior knowledge, subject to constraints such as robustness to noise.
- Application of our cell type inference approach to single-cell RNAseq data.
We encourage discussing potential projects in more detail in person. Projects can be adapted to the particular interests and skills of prospective students. Projects would consist primarily of developing approaches in Matlab (and to a lesser extent, R). Projects are open to students from both experimental and theoretical backgrounds but willingness to learn and apply programming in these environments is essential.
References
[1] Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum (Bendall et al., 2011)
[2] Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE (Qiu et al., 2011)
[3] Visualizing high-dimensional data using t-SNE (van der Maaten and Hinton, 2008)
Contact person: Manfred Claassen