IMSB News
Kian Bigovic Villi starts his PhD at IMSB
Kian started his PhD in early January 2025, supervised by Julian Trouillon and Uwe Sauer. He will be working on the effects of post-translational modifications on transcription factor activity. Welcome back Kian, and all the best!
Ruedi Aebersold was honoured with the HUPO Pinnacle Lifetime Fellowship
Congratulations Ruedi!
EUPA Juan-Pablo Albar (JPA) Proteome Pioneer Award for Pedro Beltrao

Pedro received this award for his leading and long-standing involvement in promoting the principles of sharing and integrating resources for the development of excellent research in the Proteomics field.
SNF Ambizione Grant for Julian Trouillon

Julian obtained a competitive Ambizione grant from the Swiss National Science Foundation that will enable him to start his own lab on transcriptional regulation at IMSB for the next 4 years. Congrats Julian!
Pedro Beltrao has been elected as EMBO Member

EMBO is an organization of leading researchers that promotes excellence in the life sciences in Europe and beyond. It announced the election of 120 researchers, one of which is Pedro. Through this lifelong honour, EMBO Members and Associated Members are recognized for their outstanding achievements in the life sciences.
Master/Semester computational project: Predicting transcription factors input signals using network analysis
In this project, we aim at inferring input signals affecting transcription factors activity using computational analysis of regulatory and metabolic networks.
Ruedi Aebersold and Matthias Mann receive the Dr H.P. Heineken Prize for Biochemistry and Biophysics 2024

Ruedi Aebersold, the founder of IMSB, was awarded the prestigious Dr. H.P. Heineken Prize for Biochemistry and Biophysics. He receives it together with Matthias Mann (MPI Munich) "... because they pioneered the field of proteomics and revolutionized the way in which scientists study proteins today".
Mass Spectrometry in the Life Sciences Award 2024 to Nicola Zamboni

The prize by the German Society of Mass Spectrometry (DGMS) honors outstanding method developments in and applications of mass spectrometry in the life sciences.
Sauer Zamboni group achieves LEAF sustainability certification!

The group was one of 19 labs across ETH Zurich to be certified in the LEAF pilot project.
Congratulations to Stephan Durot for passing his PhD defense
Stephan's thesis pivots on the integrated analysis of endothelial cells from different vascular beds.
Adriano Rutz wins the Gold Trophy of the National ORD Prize 2023! Well done!

The prize is awarded by the Swiss Academies of Arts and Sciences in recognition of innovative and impactful practices in the field of Open Research Data (ORD).
We welcome Jonas Runde as a new PhD student in the lab!
Jonas started his PhD in the group in December, working on protein phosphorylation and post-translational modifications in E. coli. Welcome back Jonas, and viel Erfolg!
Tomek Diederen defends his PhD Thesis
We congratulate Tomek for successfully defending his thesis on "Simulation-based metabolic flux inference" which, by his own words, all boils down to clouds vs. points. Well done!
Nicola Zamboni receives Credit Suisse Award for Best Teaching

The winner is selected by the ETH Student association to honor a individual that stands out for promoting excellence in teaching. Congratulations!
Adriano and Vince won a Poster Prizes at the annual Meeting of the Swiss Metabolomics Society
Congratulations! PS: No, Nicola wasn't involved in the selection :-)
12 new MSc students join systems biology major

We are glad to welcome new students with the beginning of the Academic Year. Good luck!
The Swiss Chemical Society honors Uwe Sauer with Simon-Widmer Award

The SCS Simon-Widmer Award honors distinguished scientists for their contribution to fundamental and applied analytical science and the education of analytical scientists.
We welcome Vilhelmiina Haavisto as a PhD student in the Sauer lab!
Vilhelmiina starts in January to work on microbial interactions in seawater communities. Good luck Vilhelmiina!
Best poster award to Daniela Ledezma!

Daniela won the best poster award at the "EMBO:Multiomics to Mechanisms symposium." Congratulations Daniela!
Congratulations to Evgeniya Warmer for successful defence of her PhD thesis!

Evgeniya successfully denfended her PhD thesis on phosphoregulation in coordinating E. coli metabolism. Congratulations Evgeniya!
Congratulations to Karin Meier for successful defence of her PhD thesis!

Karin successfully defended her PhD thesis on "Microbial metabolism along the digestive tract and identification of regulatory mechanisms". Congratulations Karin!
Farewell of Marie-Therese Mackmull

Today we say goodbye to Marie-Therese Mackmull after a successful PostDoc. We will miss you and wish you the best of luck with your future endeavors!
D-BIOL Symposium
At the D-BIOL Symposium we were able to share our research with the rest of the department. We congratulate Tetiana Serdiuk, Christian Dörig, Cathy Marulli and Franziska Elsässer on their amazing posters!
Congratulations to Monika Pepelnjak!

Monika Pepelnjak successfully defended her PhD thesis today! Congratulations Monika, we are excited to celebrate this milestone with you.
Dina Schuster talks at the ASMS Conference

At the 70th ASMS Conference on Mass Spectrometry and Allied Topics, Dina Schuster presented her project "Embracing chaos: studying flexible domains on membrane adenylyl cyclases with structural proteomics". Congratulations on your successful talk!
We welcome Sabrina Keller
We welcome back Sabrina Keller for her PhD project in the group of Matthias Gstaiger!
Best Talk award to Evgeniya Schastnaya

Congrats Evgeniya for winning the best talk award at the 4th International conference on post-translational modifications in Bacteria (https://www.sysbio.se/ptmbact2020/).
Maria Bikaki gives talk at LS2 Annual Meeting

Maria Bikaki gave an intesting talk at the LS2 annual meeting on her project "Structure determination of protein-RNA complexes of SARS-CoV-2 by Cross-Linking of Isotope-labelled RNA and tandem Mass Spectrometry (CLIR-MS)". Congratulations!
Izabela Smok joins Leitner Group for her PhD
Welcome to Izabela Smok who joined the group of Alexander Leitner for her PhD project. We are excited to work with you!
Tetiana Serdiuk gives talk at AD/PD 2022 Conference

Our PostDoc Tetiana Serdiuk gave a talk at the AD/PD Conference on "Linking alpha-synuclein aggregation and mitochondria". Congratualtions on your successful talk!
Welcome to the Slack Group

Prof. Emma Wetter Slack and her Food Immunology team joined IMSB
Welcome to Anna Pagotto

We welcome Anna Pagotto as a visiting scientist from the Univeristy of Padova. We are excited to work with you for the next six months!
Congratulations to Patrick Stalder!

We are excited to announce that Patrik Stalder successfully defended his PhD thesis today! Congratulations on the great defense!
Warm welcome to the Beltrao Group

The Beltrao Group has officially joined IMSB as of January 2022.
Viviane Reber presents at EUbOPEN Annual Meeting

At the EUbOPEN Annual Meeting 2021, Viviane Reber presented her project. Congratulations on your successful talk!
Our paper on our R package protti is finally out!
Jan-Philipp Quast and Dina Schuster published their R package protti that they developed to analyze proteomics data. Congratulations on the amazing work!
Welcome to Luca Riermeier
We welcome Luca Riermeier as the newest PhD student in the groupf of Alexander Leitner. We are excited to work with you!
Open Postdoctoral Positions in the Zamboni lab
For people passionate about metabolomics, mass spectrometry, and computational challenges.
Best poster award to Daniela Ledezma!

Daniela won the best poster award at the EMBO:Multiomics to Mechanisms symposium (https://blogs.embl.org/events/2021/10/14/multiomics-to-mechanisms-best-poster-awards/). Congratulations Daniela!
Franziska Elsässer wins Best Presentation Prize

At the LS2 Swiss Proteomics Meeting in Montreux, Cathy Marulli and Franziska Elsässer presented their projects, and Franziska even won the Best Presentation Prize. Congratulations on your successful talks!
Best poster prize!

Chris Gruber won the best poster prize at the first in person conference (Quantitative Modeling of Cell Metabolism in Kopenhagen) this year. Congrats Chris!
Jan-Philipp Quast talks at the Basel Computational Biology Conference

Jan-Philipp Quast presented the R package protti which he developed together with Dina Schuster at the Basel Computational Biology Conference. We are excited to share our work with the research community!
Master/Semestert Project: Metabolome:Proteome Cross-Talk in Basal Autophagy
For students of D-BIOL/D-HEST/D-BSSE
Master/Semester Project: Complexity of Mitochondrial Transporters in KRAS-driven Colorectal Cancer
For students in D-BIOL/D-HEST/D-BSSE
Master/Semester Project: Complexity of mitochondrial transporters and metabolic rewiring III
For students of D-BIOL/D-HEST/D-BSSE
Master/Semester Project: Complexity of mitochondrial transporters and metabolic rewiring II
For students of D-BIOL/D-HEST/D-BSSE
Master/Semester Project: Complexity of mitochondrial transporters and metabolic rewiring I
For students of D-BIOL/D-HEST/D-BSSE
Master/Semester Project: Interfacing microfluidics and high-resolution mass spectrometry for the study of complex metabolomes II
For students of D-BSSE/D-MATL/D-CHAB/D-BIOL
Master/Semester Project: Master’s Student Project: Interfacing microfluidics and high-resolution mass spectrometry for the study of complex metabolomes I
For students of D-BSSE/D-MATL/D-CHAB/D-BIOL
An all-inclusive, turnkey software for analysis of untargeted LC-MS data
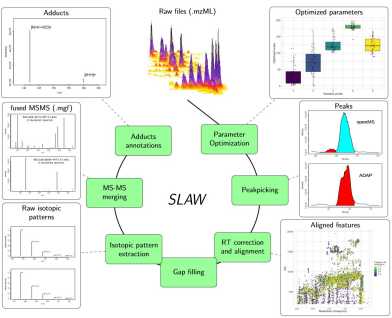
We are proud to publicly announce SLAW.
We are hiring!
We are seeking for ambitioned PostDocs in the area of mass spectronetry (metabolomics and lipidomics) and computational biology or chemistry
Paper on genome design principles published

In our Nature Communications paper, we report new genome design principles and uncovered new genetic control elements through a detailed RNASeq analysis of our rewritten C.ETH 2.0 genome. Congratulation to Mariëlle and everyone involved!
Welcome to Viviane Reber

We are excited to welcome Viviane Reber back as a PhD student in the group of Matthias Gstaiger. We wish you a successful start!
Pedro Beltrao joins IMSB

Starting January 2022, Pedro will join IMSB as a Professor for Computational Biology. We are all exited...
Zamboni lab guests Dr. Krystina Kupcova
We are happy to announce that Dr. Krystina Kupkova will start working with us from March!
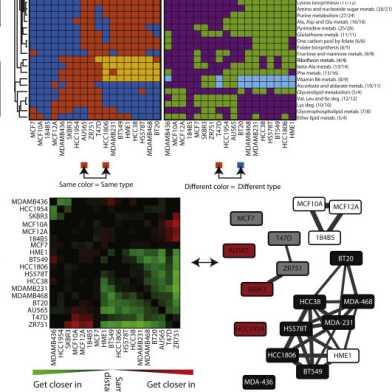
Our collaborative paper on modeling signaling networks is published.
Our paper "Deciphering the signaling network of breast cancer improves drug sensitivity prediction" is out in Cell Systems.
The Sauer group welcomes Julian Trouillon to his new post-doc position

Julian recently joined the Sauer group and will be developing experimental approaches for systematic determination of metabolite inputs into transcriptional regulatory networks and characterize the regulatory programs dictating metabolic shifts.
Sarah Cherkaoui defended her PhD at the Zamboni lab, congratulations!
In December Sarah Cherkaoui defended her thesis!
Congratulations to Philipp Warmer for successful defence of his PhD thesis!

Philipp succesfully defended his PhD thesis titled: "Towards connecting membrane organisation and cellular homeostasis in prokaryotes by lipidomics". Congratulations to Philipp and wishes of success in future challenges.
Our paper on proteome-wide structure-function is published.
Our paper "Dynamic 3D proteomes reveal protein functional alterations at high resolution in situ" is out in Cell.
IMSB Xmas in times of Corona
A magical Xmas event with the World Vice Champion in mental magic under strict COVID safety measures …..
Welcome back Cathy!
We are exctied to welcome back Cathy Marulli for her PhD project where she will work with Alexander Leitner.
Cathy Marulli wins ETH Medal

Congratulations to Cathy Marulli for being awarded the ETH medal for her outstanding master's thesis "FLiP-MS: Towards Proteome-Wide Identification of Protein-Protein Binding Interfaces and Quantification of Interface Occupancy" supervised by Christian Dörig.
Welcome to Maria Bikaki

We welcome our newest PostDoc Maria Bikaki who joined the group of Alexander Leitner. We are excited to work with you!
Jan-Philipp Quast receives grant from the Promedica Foundation

Congratuations to Jan-Philipp Quast who has received a grant from the Promedica Stiftung to fund his PhD project!
Ruedi Aebersold receives the 100th Marcel Benoist prize (highest scientific award in Switzerland (aka. the Swiss Nobel Prize))
The IMSB founding father Ruedi Aebersold was awarded the 100th Marcel Benoist prize, the highest scientific award in Switzerland, for his pioneering role in systems biology. CONGRATULATIONS!
Viviane Reber joins for Master's Thesis
We welcome Viviane Reber to the lab for her master's thesis on Parkinson's disease supervised by Tetiana Serdiuk.
A universal trade-off between growth and lag in fluctuating environments
A joint paper from the Basan, Hwa, Paulsson, and Sauer labs reports how rapid microbial growth in one condition compromises the ability to quickly switch to new conditions. This remarkably conserved tradeoff emerges directly from the biochemistry of central metabolism and can mathematically predict broadly disparate growth rates on different substrates.
Paola Picotti receives this year’s Rössler Prize
IMSB congratulates Paola Picotti on the prestigeous Rössler prize for her groundbreaking proteomics developments!
How bacteria fertilise soya
Our recent paper on symbiotic nitrogen fixation was published in Molecular Systems Biology. Read the ETH news release how bacteria fertilise soya.
Tactile sensing controls cell-cycle initiation in bacteria
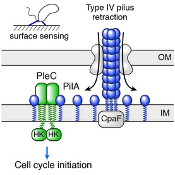
A recent PNAS paper by the Christen group shows how pili function as tactile sensors and reports on the underlying signaling mechanism that interlinks surface attachment with cell-cycle initiation in bacteria.
30 Mass Spectrometers!

IMSB reached the milestone of 30 mass spectrometers in operation! These are key for the analysis of proteins, metabolites, and lipids in complex systems. This makes IMSB one of the worldwide largest research units of this kind, and a premier site for systems biology.
New Mass Spectrometer for the Zamboni Lab

A shiny Agilent 6546 QTOF was just installed! This is part of a long-standing collaboration with Agilent.
New NCCR with IMSB participation
The IMSB labs of Paola Picotti, Uwe Sauer and Mattia Zampieri are part of the just granted National Center of Competence in Research to fight antibiotic-resistant bacteria (NCCR AntiResist). It is a collaboration with colleagues at the Biozentrum and University Hospital Basel and the ETH Department of Biosystems in Basel. The aim is to find new antibiotics and develop alternative strategies to combat antibiotic-resistant bacteria by linking basic research directly with clinical research.
Paper on field-portable bio-sensor device published.
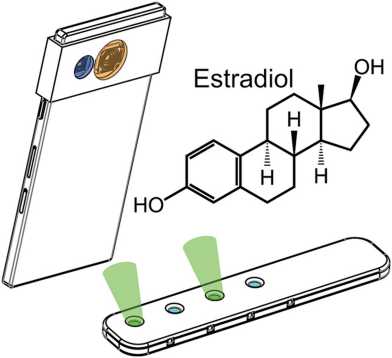
The paper, published in Biosensors and Bioelectronics, reports on a smart phone compatible biosensor-device that uses engineered yeast cells to detect endocrine-disrupting chemicals in the environment. Congratulation to Nadine Lopsiger, Jonathan Venetz and colleagues on this nice piece, which emanated from a fruitful collaboration with the group of Prof. W. Stark.
Paola Picotti receives ERC Consolidator Grant with EUR 2 Mio

Congratulations to Paola and her great team, whose amazing work in recent years made this possible!
Farewell lecture by Ruedi Aebersold

Ruedi Aebersold delivered his farewell lecture in an almost complete Audimax, entitled “Opportunities created, opportunities seized, and opportunities missed”.
Master Graduation Ceremony

At the D-BIOL master graduation ceremony five of our students were from the major in systems biology. Congratulations to all of them!
Classen group moves to Tübingen

Our non-tenure track Assistant Professor Manfred Claassen will start as a tenured Professor at the University of Tübingen (Germany) January 2020. We will miss you Manfred!
A life devoted to protein research

Ruedi Aebersold on his 30-years long journey dedicated to advancing protein research.
PHRT Hub for Clinical Mass Spectrometry
The Zamboni lab has been tasked by the PHRT EC to provide metabolomics in support of the personalized health initiative in Switzerland. Together with the PHRT Center for Proteoform Analysis, it will provide a one-stop shop for clinical mass spectrometry in Switzerland.
Patent on Parallel Functionality Testing Synthetic Genome.
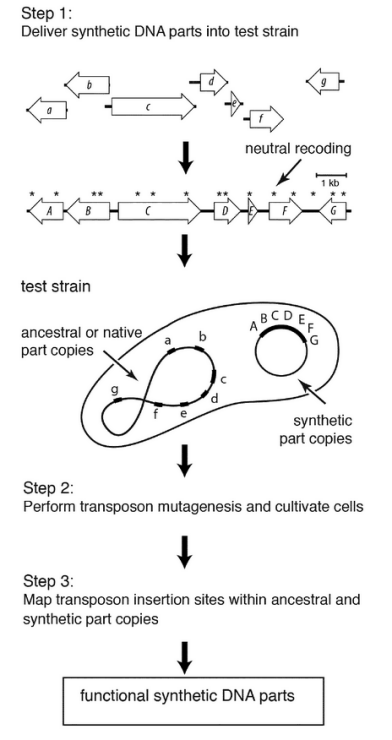
Our patent on PARALLEL FUNCTIONAL TESTING OF SYNTHETIC DNA PARTS, PATHWAYS, AND GENOMES was granted by EP and enters nationalization.
Our paper on alpha-synuclein toxicity out in Cell Systems!
Former graduate student Martin Soste studied mechanisms of genes that modulate alpha-Syn toxicity. He used the lab's "sentinel assay" to monitor the activation state of ~200 pathways in yeast and found that blocking the biosynthesis of diacylglycerol, a precursor of lipid droplets, rescued cells from toxicity. Read the paper here: https://doi.org/10.1016/j.cels.2019.07.010
Research on Computer-generated Genomes featured in NZZamS

Our research on digital genome design technology was featured in a news article in NZZ am Sonntag.
Regulatory mechanisms coordinating amino acid and glucose catabolism in E. coli
Microbes need to coordinate utilization of nutrients but the triggering signals remain unclear. A recent "Nat Comm" paper by the Zampieri & Sauer groups (IMSB) developed a computational approach to identify the intracellular signals that prioritize amino acid over glucose degradation from dynamic exo-metabolome data.
Master/semester project - Prediction and characterization of unknown membrane protein function
In this project, we will systematically characterize the function of unknown membrane proteins and their role in the cell's response to environmental conditions.
Beat Christen speaks at the World Economics Forum on Digital Genome Design Algorithms

Beat Christen discussed how computer algorithms are used to encoding new abilities into artificial DNA and the convergence between the digital and biological worlds at the WEF in Dalian. Check out the presentation on Youtube.
ETH spin-off Gigabases Switzerland AG founded

Gigabases Switzerland AG develops genome design algorithms to breed novel pathways and organisms that replace chemistry with sustainable biological processes
Leitner group news Q1/Q2 2019
A summary of the first half of 2019. Read more
Our paper on Parkinson’s disease out in Science Translational Medicine!
Former lab member Juan Gerez (now in the Riek lab) found a new ubiquitin ligase complex that targets internalized alpha-synuclein fibrils for degradation. https://stm.sciencemag.org/content/11/495/eaau6722.full
Paola wins EMBO gold medal!
Paola has been awarded the EMBO gold medal. It is awarded every year to young scientists who have made outstanding contributions to life sciences in Europe. Congratulations to Paola!
Welcome to Natalie!
Natalie de Souza joined our group in April 2019 as a Scientific Officer and Lecturer.
Welcome to Jan-Philipp!
Jan-Philipp Quast will do his PhD in our lab.
Welcome to Franziska!
Franziska Elsässer joined our lab as a PhD student in April 2019.
ETHics event at Audimax

Thanks to all participants for the engaging exchange and debate. Special thanks to VP Detlef Günther for the welcome, Prof. Yaakov Benenson, Alessandro Blasimme, Filippa Lentzos, and Prof. Kenneth Oye for sharing their expertise and insights in the panel discussion, and Prof Michael Hampe for moderating the event.
(p)ppGpp Regulates a Bacterial Nucleosidase by an Allosteric Two-Domain Switch
In a joint collaboration, researchers from Denmark and Sauer Group at IMSB have shown that bacteria produce a specific stress molecule, divide more slowly, and thus save energy when they are exposed to antibiotics. The new knowledge is expected to form the basis for development of a new type of antibiotics.
De novo Classification of Mouse B Cell Types using Surfaceome Proteotype Maps
Interested in a new way of classifying B Cell types then have a look at a recently paper from our lab which is now on BioRxiv.
First Computer-Generated Genome published in PNAS
Our work on the chemical-synthesis rewriting of a bacterial genome was published in PNAS. Congratulation to everyone involved on this achievement. Check out the genome sequence of Caulobacter ethensis 2.0, which we have physically manufactured with the help of digital genome design algorithms.
Welcome to Aleš!
We welcome Aleš Holfeld! He will be performing his PhD project in our group. Good luck!
Leitner group news Q3/Q4 2018
A summary of the second half of 2018. Read more
The protein with the starting gun
Whether dormant bacteria begin to reproduce is no accident. Rather, they are simply waiting for a clear signal from a single protein in the cell interior. The Sauer lab have now deciphered the molecular mechanisms behind this.
In silico surfaceome paper was recommended by F1000
The article "The in silico human surfaceome" has recently been published in PNAS and now has been recommended by the F1000 member Alejandro Schaffer to be of special significance in its field.
Thanatin targets the intermembrane protein complex required for lipopolysaccharide transport in Escherichia coli
Congrats to the Robinson lab and Maik Müller for getting their paper out about the molecular mechanims of action of thanatin, natural occuring antimicrobial peptide that kills Gram-negative bacteria.
We moved our lab
Together with the rest of IMSB we moved our lab to new a building, which meant 2-fold increase in office and lab space. We now got space to separate office and lab work, so we now can enjoy a coffe while doing bioinformatic analysis without atleast fearing that the coffe is contaminated. The move also meant we now got dedicated space for clinical proteomic work. You can now find us in the HPM2 building on the D floor for the clinical facilities and F floor for the research facilities.
Research paper published on chemical diguanylate cyclases modulators
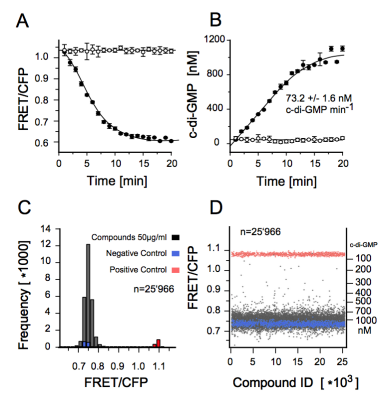
Our work on the identification of small molecule modulators of diguanylate cyclase by FRET-based high-throughput-screening was published in ChemBioChem.
Welcome back Alex
We welcome back Alex in our lab. She has been a Masters' student and now she will stay with as a research intern. Good luck, Alex!
We moved our lab!
Together with the rest of IMSB we moved our lab to a new building. For us it meant a physical move of one floor down and a few meters back, but it gave use the opportunity to put together a nice, big, open-space lab and equip it with all new instruments and items. You can find us now in HPM2, D24. Our mass specs joined the mass specs of the other IMSB groups in a massive underground lab. Watch some pictures of the progress
In silico human surfaceome published!
Congrats to the Wollscheid lab to get their paper on predicting cell surface proteins published in PNAS!
Monika awarded the ETH Medal for Outstanding Master Theses
Congratulation to Monika! She was awarded the ETH Medal for Outstanding Master Theses for the work she did in our lab as a Masters' student. Way to go!
Review about Surfaceome Nanoscale Organization now online!
Surfaceome nanoscale organisation and its extracellular interaction network determines cellular identity and fate. This review discusses different surfaceome models and which factors incluence their organisation.
Ilaria wins Early Career Researcher competition!
Big congratulation to Ilaria for winning the Early Career Researcher competion 'highlight of the year'!
Several new publications by the Wollscheid lab
The Wollscheid lab has been successful in publishing some long-standing studies: 1. The HATRIC paper 2. Proteomic profiling of poxvirus assembly 3. An antibiotic interactions study in pseudomonas aeruginosa
Welcome two new PhD student to the Wollscheid group!
Jonas Albinus (from Denmark) and Sebastian Steiner (from Switzerland) just started as PhD students in the Wollscheid lab. They will be working on clinical proteomics and we wish them a good start!
DIA/SWATH tutorial published in MSB
Our tutorial article describing everything you need to know to get started with DIA is now (finally!) published.
Video lectures from DIA/SWATH course are online!
The video lectures from the our week-long course focused exclusively on DIA/SWATH are now online.
ChristenLab hosts first iSeq instrument in Switzerland

The Christenlab received the first bench-top iSeq DNA sequencing instrument in Switzerland. The iSeq combines CMOS with on-chip sequencing by synthesis technology.
TnSeq study in Brucella published

Our TnSeq study in collaboration with Xavier DeBolle was published in Infect. Immun.
Welcome to Christian
A big welcome to Christian Dörig who is starting in our group as a PhD student. All the best for your project!
Welcome to Marie
We welcome Marie-Therese Mackmull to our lab as a post-doctoral fellow!
Leitner group news Q1/Q2 2018
A summary of the first half of 2018. Read more
Prizes for Paola and Ilaria at EuPA 2018

Big congratulations to Paola and Ilaria as they both won a prize at this year's EuPA.
PanCancer blood proteome analysis published in Cell Reports!
The article present biomarker study in blood samples in 5 solid carcinomas. Congratulations to Tatjana Sajic on positive outcome of this major effort!
Absolute stoichiometry determination of dynamic protein complexes
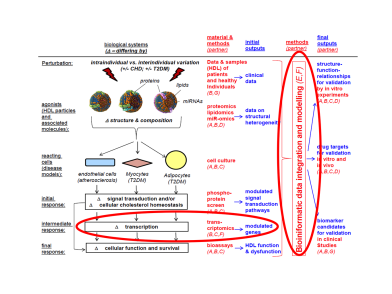
Mass spectrometry is the widely used accurate quantification of the proteome composition. However very few attempts have been made to use these techniques for the determination of the absolute stoichiometry of protein complexes. The goal of this project is to use Single Reaction Monitoring (SRM)-MS, in combination with AQUA peptides, to define the absolute stoichiometry of dynamically assembling protein complexes that are relevant in the context of inflammation and autoimmune diseases.
New students at Aebersold group!
Luzia Stalder, Chiara Auwerx, Fabian Radler and Varun Sharma have joined the group. Welcome!
Sauer-Picotti collaboration results in publication in Sci Transl Med
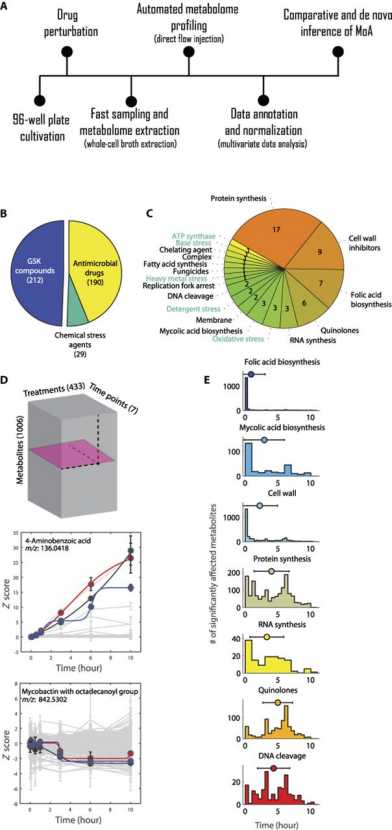
Congrats to Ilaria and Paola as their collaboration with the Sauer group and groups in Basel and Paris led to a paper in Science Translational Medicine: High-throughput metabolomic analysis predicts mode of action of uncharacterized antimicrobial compounds.
Our Cell Paper on Protein-Metabolite Interactions Recommended in F1000, twice

We thank Drs. Kuroda and Papp for recommending our article "A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication" in Faculty of 1000.
Paper using LiP for identification of protein-metabolite interactions now out in Cell!
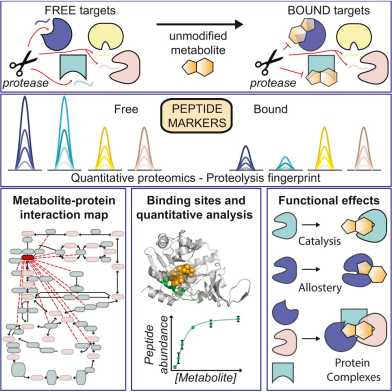
Congratulations to Ilaria, Valentina, Paola and members of the Sauer lab as their paper 'A Map of Protein-Metabolite Interactions Reveals Principles of Chemical Communication' was accepted by Cell and is now online. The paper presents a chemoproteomic workflow for the systematic identification of metabolite-protein interactions directly in their native environment.
Leitner group news Q3/Q4 2017
A summary of the second half of the year. Read more
Friedrich-Miescher-Award for Paola

We just received the exciting news that Paola will get the 2018 Friedrich-Miescher-Award from the Molecular and Cellular Biosciences section of Life Sciences Switzerland. Congratulations to her!
Claassen lab: PostDoctoral position in computational single cell biology of auto-immune diseases
A position is open for a postdoctoral research assistant in the Computational Biology Group at the Institute of Molecular Systems Biology at ETH Zürich. The available position is offered in Claassen group that focuses on concepts from statistics, machine learning and mathematical optimization to describe biological systems from single cell data. We are looking for you as of now, or upon agreement, as a
Our SPHN/PHRT Driver project PRECISE got approved!
Identification of biomarkers and therapeutic targets in inflammatory disease immunotherapy by high-dimensional single cell analysis and cluster proteomics
Joint proposal on lympathic endothelial cells approved by SNF Sinergia!

The project is a collaboration with the group of Tanya Petrova (University Lausanne) and Pia Ostergaard (St. George’s Hospital and University of London, UK). We aim at advancing our understanding of fundamental biological mechanisms driving formation and function of lymphatic vasculature in normal and pathological conditions.
Personalized health: several IMSB proposals funded

Seven projects co-submitted by IMSB members were approved in the 1st joint PHRT-SPHN call. >4 Mio CHF will flow to our Institute to develop, apply, and promote innovative technologies and platforms for personalized health.
Nationaler Zukunftstag @ D-BIOL
The Christen and Wollscheid labs hosted the "Zukunftstag @ IMSB this year
"Personalized Swiss Sepsis Study" selected as a driver project by the SPHN and PHRT!

We are delighted to announce that the PSSS consortium was selected by the SPHN and PHRT panels for funding (>5 Mio CHF). The Swiss-wide consortium is headed by Prof. Adrian Egli (Uni Basel) and Prof Karsten Borgwardt (ETH Zürich). The Zamboni lab is responsible for the molecular profiling of >8000 plasma samples.
The Zamboni lab receives a grant for the standardization of non-targeted metabolomics in personalized health

We are glad to announce that our SFA-PHRT technology transfer project proposal "Strategies and standards for high-throughput, long-term, non-targeted metabolome analysis of human biofluids" has been funded!
Project on metabolic engineering of the hexosamine pathway gets funded!

The joint proposal that combines synthetic Biology (Prof De Mey, Ghent University) and systems biology (Zamboni lab) has been just approved by the FWO/SNF. Looking forward to joining forces!
Master thesis in Wollscheid Lab
Mass Spectrometric Analysis of Hepatitis B Virus X Proteoforms and Interactomes.
Nature Protocols paper describing Limited Proteolysis coupled to MS now out

Congrats to almost all of us: the protocol paper in which we practically describe the method of coupling limited proteolysis to mass spectrometry is now out in Nature Protocols! This will be an important starting point for external labs to try out this exciting and powerful method.
Welcome to Patrick and Monika
A warm welcome to Patrick Stalder and Monika Pepelnjak who have started their PhD-project and fast track Master/PhD-project, respectively, in our lab.
Hafen Lab: Semester project and/or Master thesis on the genetics of inter-individual variations in human sensory perception
The project lies at the intersection of genetics, psychophysics of smell, and data democracy. Students will have the opportunity to combine wet lab work, study design, and computational analysis. Students should be able to write simple scripts in R, Matlab, or Python or have the willingness to acquire and develop these skills.
Review on Quantitative proteomics of model organisms published in COSB
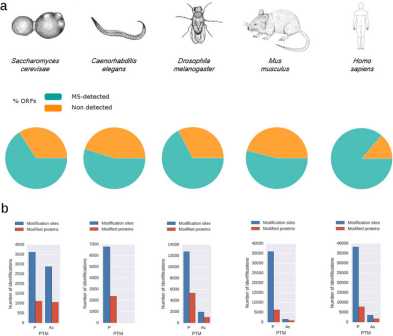
Congratulations to Yuehan, Valentina and Paola as their review on quantitative proteomics of model organisms is now published in Current Opinion in Systems Biology.
Amir's review describing proteomics approaches in Mycobacterium tuberculosis is published in COM
'Systems proteomics approaches to study bacterial pathogens: application to Mycobacterium tuberculosis' was published in an issue of Current Opinion in Microbiology focused on Bacterial Systems Biology.
The Picotti lab joins IMSB
Although they did not yet move their lab physically, the Picotti group has just changed affiliation by joining the Institute of Molecular Systems Biology of ETH Zurich, where Paola obtained a tenured position.
Mcs and Semester students join the ChristenLab

We are excited to welcome Cathy Marulli, Martin Steiner and Philipp Schächle, who start their Bachelor, Semester and Master projects in our group.
TnSeq analysis of Bacterial Methyotrophy published in Current Biology
Our joint paper with the Vorholt Lab, ETHZ was published in Current Biology.
Sauer Lab: Master thesis "Investigating protein-metabolite interactions in E. coli"
In this project, we aim at inferring functional protein-metabolite interactions in the bacterium Escherichia coli from dynamic metabolomics data.
Collaborative effort published in Nature Cell Biology

Congratulations to Yuehan and Paola as there collaborative effort with members of the Matthias Peter lab was published in Nature Cell Biology!
Paper outlining best practice in error rate control in DIA/SWATH studies published
In this study we investigated the the effect of varying library size and sample number on the accumulation of false positives in DIA/SWATH data and outlined strategies to control the error rate appropriately.
Study comparing performance of SWATH-MS across labs is published in Nature Communications
In this project led by researchers at ETH Zurich and Sciex we demonstrated the similarity of quantitative proteomics data generated by 11 labs worldwide from benchmarking samples using SWATH-MS.
Hafen lab: Master’s thesis and/or semester project on the role of cryptic genetic variation on circadian behaviors
Cryptic genetic variation (CGV) is the hidden genetic variation accumulated over time that do not contribute to the normal range of phenotypes, but may provide a source of adaptive variation to novel conditions. This project will examine if the CGV in different genetic backgrounds is influential in the evolution of circadian traits. (Keywords: Drosophila melanogaster, genetics, behavior, circadian rhythms)
Charlottes's review on host-pathogen protein interactions is published in COM
'Elucidation of host–pathogen protein–protein interactions to uncover mechanisms of host cell rewiring' was published in an issue of Current Opinion in Microbiology focused on Bacterial Systems Biology.
F1000 Research poster award at ISMB/ECCB 2017 in Prague for Jelena Čuklina
Jelena Čuklina has received a F1000 Poster Award for the work entitled "Selection of Stable Biomarker Signature for Prediction of Metabolic Phenotypes". Congratulations!
Videos from the first DIA/SWATH course available
The video lectures from the first week-long course to be focused exclusively on DIA/SWATH are now online.
TnSeq study uncovers mechanism of evolvability within a bacterial pathogen
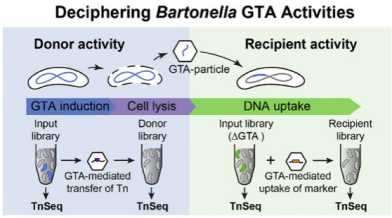
Our transposon sequencing analysis of the gene transfer system in Bartonella henselae showed that genetic exchange preferencially occures among the fittest subpopulation of cells. Our joint paper with Christoph Dehio, University of Basel, was published in Cell Systems.
Our computational platform to accelerate genome synthesis is published in PLoS One.

We present the Genome Partitioner, a broadly applicable DNA design platform, to construct genome-scale DNA programs using optimized assembly protocols according to retro-synthetic principles.
Hafen Lab: Master thesis/semester project on CRISPR/Cas9-mediated SNP changes to elucidate cause of size variations
How genetic variation (e.g. SNP) leads to different traits/phenotypes in natural populations is a fundamental and challenging problem in both the life and medical sciences. This project will involve several objectives to tackle this problem by first establishing an efficient in vivo CRISPR/Cas9 method and then using this method to change phenotype-associated SNPs in inbred Drosophila lines derived from a natural population.
Professor Ruedi Aebersold awarded Barry L. Karger Medal
Prof. Ruedi Aebersold has been awarded Barry L. Karger Medal for significant contribution to the development of new bioanalytical methods. Congratulations!
Three (!) IMSB students win ETH Prize for oustanding doctoral thesis
We are honored and proud to announce that George Rosenberger (Aebersold lab), Daniel Sévin (Sauer lab), and Andreas Kühne (Zamboni lab) won the prestigious ETH Silver Medal for Oustanding Doctoral Theses. Congratulations!
Our Science paper discussed in Quanta Magazine
Our recent paper "Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability" (Leuenberger et al. Science 2017) is discussed in the Quanta Magazine.
Johannes' paper on HIV infection proteomics published in MCP
'In Vivo and in Vitro Proteome Analysis of Human Immunodeficiency Virus (HIV)-1-infected, Human CD4+ T Cells' published in a special issue of MCP focused on infectious disease.
Faculty of 1000 recommendation for our Science paper

Our recent Science paper was recommended by faculty members at Faculty of 1000.
Highlight in BioTechniques

Our recent paper "Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability" (Leuenberger et al. Science 2017) is highlighted in BioTechniques.
HFSP scholarship won by Duncan Holbrook-Smith
Duncan Holbrook-Smith won a prestigious HFSP scholarship for his postdoc project at the Sauer lab, on exploiting non-targeted metabolomics for parallelized discovery of chemical probes for receptor-like proteins, and using those chemical probes to better understand the specific roles of receptor-like proteins in that system. Now he has the problem of deciding which fellowship to accept………
Welcome Jonathan Venetz

Jonathan joined the Christen lab for his PhD.
Article in Neue Zürcher Zeitung about our paper in Science

The Neue Zürcher Zeitung (the Swiss newspaper of record) has published an article about our recent paper in Science.
Our Science paper on proteome thermostability highlighted in ETH News
Our paper on the study of the determinants of proteome thermostability that was just published in Science has been highlighted in the ETH News.
Our study on the determinants of proteome thermostability was just published in Science!

Our paper entitled "Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability" has just been published in Science.
Eirini's paper accepted in Nature Communications
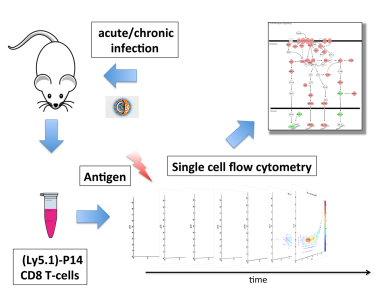
Sensitive detection of rare disease-associated cell subsets via representation learning. Using CellCnn, we identify paracrine signaling-, AIDS onset- and rare CMV infection-associated cell subsets in peripheral blood, and extremely rare leukemic blast populations in minimal residual disease-like situations with frequencies as low as 0.01%.
Welcome Kathrin Frey
After completing already her semester project in the Wollscheid lab, Kathrin now started her Master thesis investigating the uptake and the involved signalling pathway of HDL particles.
Michael Zimmermann and Paul Murima won the Swiss TB 2017 Award!
Michael Zimmermann and Paul Murima won the Swiss TB 2017 Award for their Nature Communications paper "A rheostat mechanism governs the bifurcation of carbon flux in mycobacteria." CONGRATULATIONS!
Genome-wide landscape of gene-metabolite interactions charted for bacterium
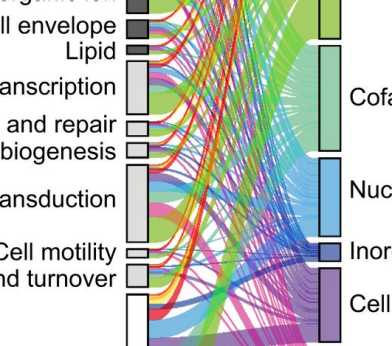
How do gene (knock-outs) affect metabolite levels? We tested this systematically for > 3'800 genes in E. coli and found several suprises.
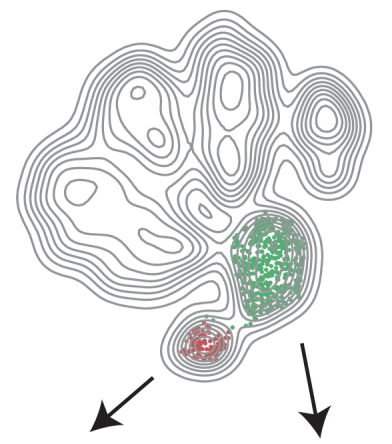
Metabotypes of breast cancer cell lines revealed by non-targeted metabolomics
A metabolome and network centric analysis of 18 breast cancer cell lines reveals a fine-grained metabolic heterogeneity. The study will appear in a special issue of Metabolic Engineering devoted to cancer metabolism. We thank the team of our collaborator Prof. Almut Schulze (Würzburg).
Nontargeted in vitro metabolomics for high-throughput identification of novel enzymes in Escherichia coli
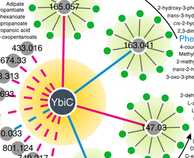
About 15'000 high-throughput, non-targeted metabolomics analyses were acquired to study >1'200 functionally uncharacterized proteins. The study was just published by Daniel Sévin and co. in Nature Methods.
Sauer group: Interplay of posttranslational modification sites in enzyme activity regulation (Master project)
Research project available on investigating the interplay and protein activity fine-tuning of different posttranslational modifications (PTM) sites
Visit of Emily LeProust at ETH

The Christen lab is proud to host Emily LeProust, CEO of Twist Biosciences during her visit at ETH.
Sauer group: Metabolomics and Flux Balance Analysis in Saccharomyces cerevisiae (Semester/Master Project)
Research project available on investigating the regulation of metabolism in S. cerevisiae, via enzyme phosphorylation
Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals
Histone modification profiling in rare stem cell populations: our collaborative work on the epigenetics in muscle stem cell aging got published in Nature!
Anna's paper accepted in PLOS Computational Biology.
Sparse Regression Based Structure Learning of Stochastic Reaction Networks from Single Cell Snapshot Time Series
Justins' paper came out in Cell Systems
Analysis of Cell Lineage Trees by Exact Bayesian Inference Identifies Negative Autoregulation of Nanog in Mouse Embryonic Stem Cells
Future Day at ETH

The Christen Group hosted a group of 15 children during the ETH future day.
Welcome Mariëlle Van Kooten
Mariëlle joined the Christen lab for her PhD.
Mcs and semester students join the ChristenLab
We welcome Donat Appert and Alexander Wölfle, who started their master and semester projects in the ChristenLab.
Arginine Modulates T Cell Metabolism and Enhances Survival and Anti-tumor Activity
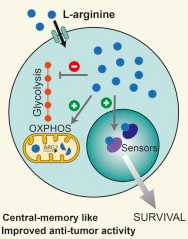
We are proud to announce that our joint study with prestigious colleagues of IRB Bellinzona, ETH Zurich, MPI Martinsried finally published in Cell: from omics to mechanism and anti-tumor effect in mice!
Welcome Fabian Wendt
Fabian studied Biochemistry at the Ruhr-University in Bochum, Germany. He started recently as PhD student in the Wollscheid Group and will drive the development of the ligand-receptor capture (LRC) technology towards new application spaces and the characterization of dynamic protein-protein interactions.
SUMOFLUX paper published!
We congratulate Maria for publishing her novel method for targeted 13C flux ratio analysis.
Welcome to Valentina and Thomas
Welcome (back) to Valentina Cappelletti and Thomas Hauser. They will start their post-doc projects in our lab from today onwards. Good luck!
Zamboni group, Metabolomics & network analysis
Research project available on investigating metabolic response to mitochndrial defects in yeast
Lund University awards honorary doctorate to Prof. Ruedi Aebersold

Great honour for Prof. Ruedi Aebersold: The Faculty of Medicine of the Lund University has awarded the honorary doctorate to him.
Annie Champagne Queloz defended her PhD Thesis
The subject of her work was: " Biological thinking: insights into the misconceptions in biology maintained by Gymnasium students and undergraduates” Congrats Annie!
Welcome Lili!
We welcome Lili Malinovska who will join our lab as a Post-doc. All the best for your project!
Uwe Sauer newly elected EMBO Member

Congratulations!
A Genome-Scale Database and Reconstruction of Caenorhabditis elegans Metabolism published
We congratulate Prof. Christoph Kaleta (Jena), his team, and all collaborators for their achievement!
Cell cycle coordinates carbohydrate metabolism in yeast
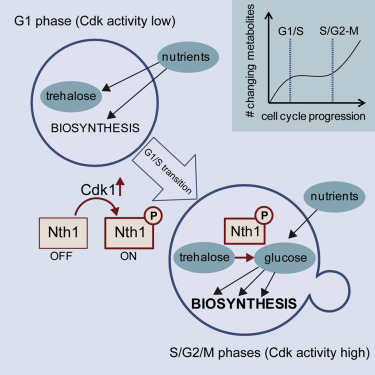
We are proud to announce that our paper on The yeast cyclin-dependent kinase routes carbon fluxes to fuel cell cycle progression was published today in Molecular Cell. The study was conducted jointly with Jenny Ewald and Jan Skotheim (Stanford).
Ketogenic diet rescues mouse embryos missing the mitochondrial pyruvate carrier
Our SNF Sinargia work with the Martinou Lab (U. Geneva) was published in PLoS Genetics. Congratulation to his team, Petra and Manuel!
Aebersold group: Student project to monitor epigenetic states in melanoma cells using novel proteomic tools
The student will learn how to process samples for mass spectrometry analysis, to analyse the resulting proteomic datasets and integrate them into a larger multi-omics framework. The work will be published in a scientific journal and the student will be involved in writing up the manuscript.
Christian Feller obtained grant for melanoma epigenetic profiling
Christian obtained funding from the 3rd URPP UZH Translational Cancer Research call to monitor epigenetic states in melanoma cells. For this project, we are currently looking for MSc students and students helpers.
Master thesis in Wollscheid Lab
Mass Spectrometric Analysis of Hepatitis B Virus X Proteoforms and Interactomes.
Andreas Kühne defended his PhD thesis
The subject of his massive work was "Development and application of graph-based computational methods to elucidate the functional role of metabolism in human skin". Congratulations!
ChristenLab, Block-course in Synthetic Genomics

Welcome Alexander, Florin and Timon to the Synthetic Genomics Block-Course offered by the Christen Group. Students will obtain training in large-scale DNA assembly techniques and gain insights into computational approaches for bio-systems design.
Charlotte Nicod joins the group as PhD student
Charlotte will work on detecting host-pathogen protein-protein interactions (HP-PPI) in Mycobaterium tubercuosis
Daniel Sévin defended his PhD thesis
We congratulate Daniel for his work on "Nontargeted metabolomics appraoches for enzyme discovery and high-resolution metabolome profiling"
Martin awarded silver medal for "Outstanding Doctoral Theses"

Martin was awarded the Silver Medal of ETH for “Outstanding Doctoral Theses”. That means that Martin’s thesis was among the top 5% PhD theses awarded by ETH each year, based on a jury of international reviewers. Btw, there are no gold or bronze medals.
Paola winner of the 2016 SGMS award!

We are proud to announce that Paola has won another prize: the Swiss Group for Mass Spectrometry (SGMS) award.
The maternal microbiota drives early postnatal innate immune development
In collaboration with the Sauer lab, Mercedes Gomez de Agüero and Stephanie C. Ganal-Vonarburg from the Maurice Müller Laboratories (DKF), University of Bern, describe in detail how in gestating mice, the mother’s community of microbes shapes the immune system of the offspring.
Stocker lab, Master Thesis Project on impact of amino acids on tumor growth
Investigate the influence of amino acids on the hyperproliferative potential of PTEN mutant cell populations in Drosophila melanogaster.
F1000 award at the Life Sciences PostDoc day 2015 for Christian Feller
Christian Feller receives F1000 award at the Life Sciences PostDoc day 2015 For his talk entitled “Dissecting protein modification-mediated communication”. Congratulations!
Donation of Lotte and Adolf Hotz-Sprenger Foundation for Prostate Cancer Digital Biobanking
With its generous donation to the ETH Zurich Foundation Lotte and Adolf Hotz-Sprenger Foundation supports the project "Prostate Cancer Digital biobanking" within the "Personalized Medicine" initiative of ETH Zurich, the University Hospital Zurich and the University of Zurich.
Nature Biotechnology asked Paola's opinion, this is what she said

Nature Biotechnology asked a selection of researchers about the most exciting frontier in their field and the most needed technologies for advancing knowledge and applications. Also Paola was asked for her opinion.
Paola wins Robert J. Cotter New Investigator Award 2016

We're to proud to have learned that Paola has won the 2016 Robert J. Cotter new investigator award for her significant achievements in proteomics.
Claassen Lab Semester Project: Deep Learning for dynamic system motif detection in health and disease
Cells process external and internal biochemical signals by means of signaling cascades, which constitute complex, dynamical networks. In this project we use novel deep (machine) learning techniques to decompose such networks in their building blocks in order to elucidate the overall structure.
Christen Lab: Master Thesis Project in Experimental Systems Biology
Discovery of genetic redundancies using a targeted genome reduction approach
Wollscheid Lab, Master Thesis in Receptor Nanoscale Organisation & Cellular Signaling
The research of our lab focuses on cell surface protein in general and specifically on receptors. We recently suceeded in developing a technology which enables the decoding of ligand-receptor interactions [1].
Just out: two papers we contributed to.
Paola, Andre and Juan contributed to two projects of which the results were recently published in Journal of Biological Chemistry and PLoS Genetics.
Paper on quantitative selection analysis featured in JMB

Our latest paper on quantiative selection analysis of bacteriophage susceptibility in Caulobacter crescentus was published as a featured article in the Journal of Molecular Biology.
SCNAT Rigi workshop on Systems and Synthetic Biology of Microbes

The SCNAT workshop on advances in microbial systems and synthetic biology toke place January 24th to 26th at the beautiful summit of Rigi.
Hafen Group: Functional Validation of Genes Associated with Starvation Resistance
Hafen Group Student Project
EMBO fellowship for Valentina

Congrats to Valentina Cappelletti who received a EMBO short term fellowship to perform part of her PhD project in our lab!
Yuehan and Paola wrote a practical guide to measuring proteins by SRM

As part of the Springer Protocols Methods in Molecular Biology book entitled Proteomics in Systems Biology, Yuehan and Paola wrote a practical guide about Selected Reaction Monitoring to Measure Proteins of Interest in Complex Samples.
Aebersold lab, Proteomics, Computational mass spectrometry and Immunology
Two bioinformatics projects available
Aebersold Lab, Proteomics, Method development in chemical cross-linking
Master thesis or semester project
Aebersold Lab, Proteomics, Method development in the field of phosphoproteomics
Master thesis or research projects available
Two new papers published by our collaborators
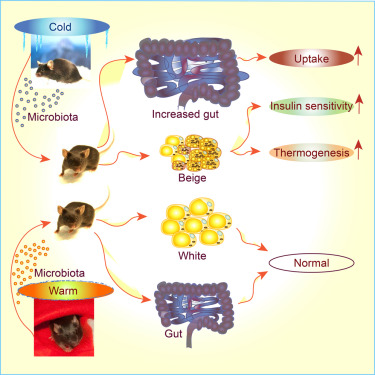
We congratulate with Prof. Trajkovski (U. Geneva) for demonstrating how "Gut Microbiota Orchestrates Energy Homeostasis during Cold" and Prof Ristow (ETH Zurich) to demonstrate the role of branched chain amino acids in regulating aging.
Ulrich Genick awarded with “Golden Owl” of the VSETH

for outstanding contributions to teaching. Congratulations!
Luca Del Medico starts his PhD
Luca performed his master thesis in our group and now joined the Christen lab for his PhD. Luca will work on novel systems and synthetic biology approaches to study core networks in prokaryotic organisms.
Pseudo-transition Analysis Identifies the Key Regulators of Dynamic Metabolic Adaptations from Steady-State Data
The Sauer Lab developed an experimental-computational approach to identify regulatory needles in metabolic haystacks. By using steady-state 13C flux, metabolites and transcript data, the approach revealed the sparse and transition-dependent regulators that drive transitions between carbon sources in Escherichia coli. Thus, pseudo-transition analysis is a novel approach to efficiently explore the vast regulatory landscape of dynamic adaptations using relatively few stationary observations.
Wollscheid Lab: Master Thesis in Receptor Nanoscale Organisation & Cellular Signaling
The research of our lab focuses on cell surface protein in general and specifically on receptors. We recently suceeded in developing a technology which enables the decoding of ligand-receptor interactions [1].
Christen Lab: New openings for Master students
Mapping cellular core functions by transposon mutagenesis and next generation sequencing
Paola named EMBO Young Investigator

Paola Picotti was selected as EMBO Young Investigator. Congratulations!
US patent issued!
Our US patent 9150916 on “Compositions and methods for identifying the essential genome of an organism” was issued on Oct. 6th 2015. The Method relates to our PCR based transposon sequencing (TnSeq) strategy to define essential DNA parts of an organism with base pair resolution.
We welcome Martin Spillmann and Donat Appert
Martin joined the Christen group for his master thesis and Donat started his semester project in our lab. Welcome on board!
Dynamic exometabolome analysis reveals active metabolic pathways in non-replicating mycobacteria
In a collaborative effort of the Sauer and Zamboni labs at IMSB and the Berry Lab at NIH, Michael and his co-workers integrated dynamic changes in the secretion and uptake of metabolites with genome-scale metabolic network reconstructions and identified active metabolic routes necessary for mycobacteria survival under hypoxic conditions.
Welcome to Valentina Cappelletti
Welcome to Valentina Cappelletti who will visit our lab for several months.
Targeted Proteomics course registration is open!
In this course we offer theoretical introductory lectures, comprehensive hands-on training and discussion rounds on each of the typical tasks performed in targeted proteomics workflows. In previous years the focus of the course has been entirely on SRM (selected reaction monitoring). This year the course content is being expanded to include other methods such as PRM (parallel reaction monitoring) and DIA (data independent acquisition), with a particular focus on SWATH MS.
Aebersold lab: Master thesis/semester project in Structural Proteomics
We are looking for motivated semester/master students to join our structural proteomics team at ETH to work at the interface between proteomics and structural biology.
Sauer Group, Metabolism, Systems Biology, Modelling, Bistability
Understanding history-dependence (hysteresis) of growth states in E. coli
Real-time metabolome profiling of the metabolic switch between starvation and growth
The Zamboni and Sauer Lab developed a platform for real-time metabolome profiling by directly injecting living bacteria, yeast or higher cell suspensions into a high-resolution time-of-flight mass spectrometer. We congratulate Hannes and all the coauthors.
Posttranslational regulation of microbial metabolism
In a review published in "Current opinion in Microbiology“, we discuss the division-of-labor between the different regulatory layers in making microbial metabolic decisions, and highlight the current challenges we are facing when elucidating the role of posttranslational regulation.
The outer mucus layer hosts a distinct intestinal microbial niche
In collaboration with the Sauer lab, Hai Li & Julien P. Limenitakis from the Maurice Müller Laboratories (DKF), University of Bern, describe in detail the compositional differences between lumen and outer mucus layer of the intestine in mice and its consequence for the present microbiota.
Cyclic operation of the pentose phosphate, Entner-Doudoroff and glycolytic pathways in Pseudomonas putida
In collaboration with the Sauer lab, Pablo Ivan Nikel from the Centro Nacional de Biotecnologia (CNB-CSIC, Spain), describes a cyclic operation of the pentose phosphate, Entner-Doudoroff and glycolytic pathways in Pseudomonas putida.
Aebersold lab, student (master) project in phosphopeptide enrichment.
We are looking for students who would like to join project for development of the new phosphopeptide enrichent for the analysis of small sample amounts. These projects are the perfect opportunity to get hands-on experience with proteomics methods and mass spectrometry instrumentation. Prior experience with mass spectrometry is not essential, although candidates with basic knowledge of proteomics methods are preferred.
We welcome Amelia Palermo!
Amelia joined the lab as an Academic Guest from the Università La Sapienza of Rome to exploit non-targeted metabolomics in the context of doping.
Our paper on acute PPP activation featured in the press
The article was picked by the Editor in Science Signaling. A preview commentary by Dick and Ralser published in Molecular Cell.
Yvonne Bösch completed her Master
Congrats to Yvonne Bösch who completed her master thesis in the Christen lab. We are excited that Yvonne will continue working in the field of synthetic biology will soon started her internship at Evolva.
An open-source computational and data resource to analyze digital maps of immunopeptidomes
We congratulate Etienne Caron and his collaborators!
Sauer Lab, Predicting antimicrobial interactions
A project combining wet and dry bench
Welcome Maik Müller
After already completing a semester project and a master project in the Wollscheid lab, Maik now started as PhD student.
A biosensor for the activity of mitochondrial pyruvate transport
Just published in Molecular Cell. We congratulate our collaborator Jean-Claude Martinou (U. Geneva) and his team for the invention.
Sauer Group, Oxidative Stress, Metabolomics, Kinetic Modelling
A comparative study of the immediate metabolic response to oxidative stress in E. coli, Yeast and Human cells
TnSeq paper on uranium detoxification published
The TnSeq study to understand uranium tolerance of Caulobacter crescentus was published in J. Bacteriol. Special thanks goes to our collaborators Yongqin Jiao and Mimi Yung from Lawrence Livermore National Laboratory
The acute response of skin cells to oxidative stress and sun irradiation
To timely celebrate one of the hottest and sunniest Summers, the Zamboni lab published a study elucidating a mechanism that allows skin cells to immediately fight oxidative damage before long-term countermeasures are available. We congratulate Andreas, Hila, and our collaborators at Beiersdorf AG
Sauer Group, Metabolomics, Antibiotics, More of action
Identifying the target of the antimicrobial peptide Sublancin
Our bioinformatic genome refactoring approach is published in ACS Synthetic Biology

We present Genome Calligrapher, a computer-aided design web tool intended for whole genome refactoring of bacterial chromosomes for de novo DNA synthesis.
Hypoxia-induced consumption of fructose provokes heart disease
The study conducted by Prof Krek was published in Nature and demonstrates how alternative splicing mediates a metabolic switch that bypasses carbohydrate assimilation in cardiac cells. We congratulate our collaborators!
Christian Feller was selected for an EMBO Long-Term Fellowship

Congratulations!
Sauer Group, Allosteric Regulation, Metabolic Network, Kinetic Modeling
Kinetic reconstruction of E. coli central carbon metabolism with allosteric regulation
Master Thesis Project in Proteomics, Computational mass spectrometry and Immunology
We are looking for 2 highly motivated and focussed MSc students with skills in bioinformatics for immunopeptidome analysis, who will: 1) automate a computational workflow for the analysis of immunopeptidomic SWATH MS data. 2) analyze a tissue-based map of the immunopeptidome. Contact: Dr. Etienne Caron
Master Student Project in Proteomics, Epigenetics in Aging and Cancer
Epigenetic pathways are major regulators of aging and aging-associated diseases including cancer. The student will apply a recently developed mass spectrometry-based method to profile epigenetic states during aging and cancer development. Contact: Dr.Christian Feller
Review on Metabolomcs and Flux Analysis published in Molecular Cell
The vignette was contributed jointly with the colleagues Prof Gary Patti (Washington University) and Prof Alan Saghatelian (Salk Institute)
An novel method for rapid and randomized prototyping of FRET sensors for small molecules
Este Peroza published the invention in Analyst.
Causal metabolic signals and functionality of protein phosphorylation in TOR signaling identified
In two related dynamic omics studies, researchers from the Aebersold and Sauer lab identified causal metabolic signals and functionality of protein phosphorylation in TOR signaling.
Functional screening identifies MCT4 as a key regulator of breast cancer cell metabolism and survival.
The collaboration was led by Prof. Almut Schulze from the University Würzburg and published in the Journal of Pathology. Congratulations!
Claassen Group: Causality from single-cell data

Causality inference from single-cell data. Inferring cause and effect, instead of mere correlative relationships between biological variables is a challenge of interest in many biological applications. This project deals with analyzing intervention experiments with single cell readout to infer causal signaling relationships.
CSPA published in PlosONE
The Cell Surface Protein Atlas (CSPA) got published in PlosONE and is finally online.
Paper published on FRET sensor for Trehalose-6-P
We congratulate our alumnus Este and our collaborator J. Ewald from the Skotheim lab at Stanford
SNF proposal approved!
Bernd Wollscheid got a three-year funding for his proposal at the Swiss National Foundation.
Martin Soste got his PhD!

Congrats to Martin who succesfully defended his PhD.
Research Assistant/PhD student
A position is open for a research assistant/PhD student in the Computational Biology Group at the Institute of Molecular Systems Biology at ETH Zürich. The available position is offered in Claassen group that focuses on concepts from statistics, machine learning and mathematical optimization to describe biological systems from single cell data. We are looking for you as of April 15, 2015, or upon agreement, as a Research Assistant / PhD student
Research Assistant/PhD student
A position is open for a research assistant/PhD student in the Computational Biology Group at the Institute of Molecular Systems Biology at ETH Zürich. The available position is offered in Claassen group that focuses on concepts from statistics, machine learning and mathematical optimization to describe biological systems from single cell data. We are looking for you as of April 15, 2015, or upon agreement, as a Research Assistant/PhD Student
Sauer Group, Metabolomics, Screening, Physiology
Investigating tradeoffs between fast growth and fast switching in microorganisms
Sauer Group, Metabolomics, Modeling, Bistability
History-dependence (hysteresis) of growth states in E. coli
Population level variability of plasma protein abundance
Researchers from the Aebersold group used a longitudinal twin strategy and SWATH mass spectrometry to characterize the quantitative variability profile of human plasma proteome.
Proteomics review online
The review, Proteomics beyond large-scale protein expression analysis, that Paul, Abdullah and Paola wrote for Current Opinion in Biotechnology is now online.
Sauer Lab, Metabolomics, Functional genomics
Proteome-scale enzyme discovery in yeast using nontargeted metabolomics.
Welcome to Natalia and Marco
We started our new year with two new group members: Natalia Prymaczok and Marco Tognetti.
Dr. Martin Mehnert was selected for an EMBO Long-Term Fellowship

Congratulations
EMBO fellowship for Ilaria
Congratulations to Ilaria who won a EMBO long-term fellowship!
Zamboni Lab, 13C metabolic flux analysis in Mycobacteria
Apply a novel framework for 13C flux analysis to co-metabolism of mycobacteria
Opportunities and Challenges of Citizen Science Workshop (January 22-23 2015)

This workshop aims at providing an overview of the present state of citizen science, an outlook into the potential of citizen science in the future and the discussion of the opportunities and challenges associated with the re-emergence of this discipline for scientists, policy makers and funders.
Welcome Marc van Oostrum!
Marc started as new PhD student in the Wollscheid Lab.
Protein acetylation affects acetate metabolism, motility and acid stress response in E. coli

Tobias Fuhrer and Uwe Sauer lab contributed to the study lead by Manuel Cánovas (Uni. de Murcia, Spain).
The role of phosphorylation in regulating yeast metabolism

Large-scale study published by the Sauer Lab in collaboration with Christian von Meering (Uni Zurich)
Christen Lab, Experimental Systems Biology
Mapping essential core functions of microbial genomes by transposon mutagenesis and next generation sequencing
LiP-SRM paper highlighted in Nature Methods
Our limited proteolysis SRM paper was highlighted in Nature Methods.
Review on biological insights obtained by nontargeted metabolomics
We congratulate and thank Daniel and Andreas for assembling this criticial review.
Paola in the Top 40 under 40

Paola Picotti made it into The Analytical Scientist's Top 40 under 40!
Review on omics for understanding disease and aiding diagnosis online
Paola contributed to the review.
Yibo Wu receives F1000 award at the IBS-IMSB Postdoc day

For her talk entitled “Mitochondrial activity – A multilayered Omics View”. Congratulations
Perspective on Systems Biology education for graduate students.

An interview to Prof. Uwe Sauer published in the SystemsX.ch newsletter
Claassen group, Computational Systems Biology
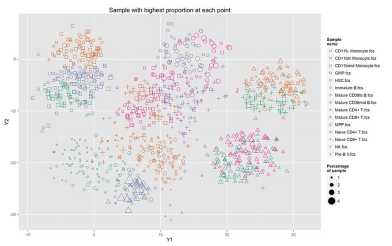
Ab initio cell type definition from high-dimensional single-cell data
Claassen group, computational systems biology
Infer structure and parameters of biochemical networks through machine learning
Paola in "The Author File" in Nature Methods

Paola was interviewed by Nature Methods on the occasion of the publication of the sentinel paper and appears in The Author File.
Sauer Group, Microbial metabolomics and modeling
Understanding the role of bacterial metabolism in the evolution of antibiotic resistance
Michi Zimmermann gloriously defended his PhD thesis
Congratulations!
Current Opinion on High-throughput metabolomics
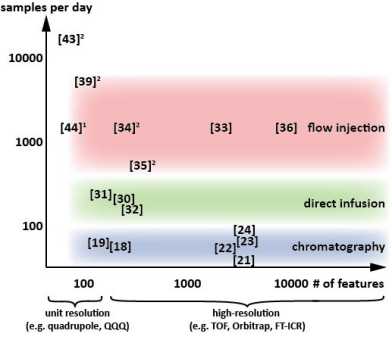
Tobias Fuhrer and Nicola Zamboni reviewed the current standing of HT metabolomics in the context of discovery.
Limited Proteolysis paper online
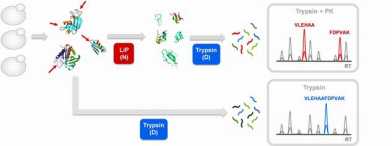
Our paper "Global analysis of protein structural changes in complex proteomes" alias the LiP paper is online on the Nature Biotechnology website.
Sentinel protein assay paper online
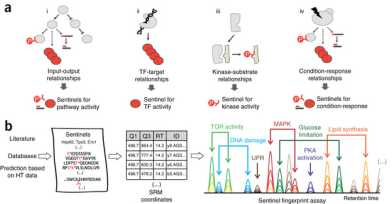
The paper "A sentinel protein assay for simultaneously quantifying cellular processes" is now available for download on the Nature Methods website.
Paper accepted in J Virol
Yuehan and Paola contributed to the paper "Stepwise priming by acidic pH and high K+ is required for efficient uncoating of influenza A virus cores after penetration" that was just accepted in Journal of Virology. Congratulations!
Paper Published
Martin and Paola contributed to the paper "Calcineurin determines toxic versus beneficial responses to α-synuclein" that is now published in PNAS. Congrats!
Discovery of Glycoprotein Biomarkers for Aggressive Prostate Cancer
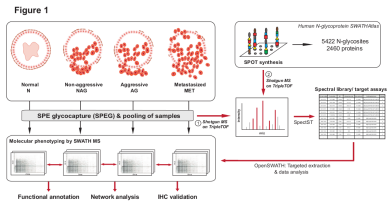
Researchers from the Aebersold group used SWATH mass spectrometry to profile the glycoproteome extracted from clinical tissues for the discovery of aggressive prostate cancer biomarkers.
Software for High-Throughput targeted analysis of SWATH-MS data
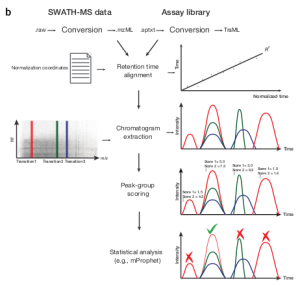
IMSB researchers describe an open-source software which allows targeted analysis of SWATH-MS data, a novel type of targeted mass spectrometry first introduced by the Aebersold lab in 2012. The OpenSWATH software allows to increase the number of analyzed peptides in targeted experiments by up to 100-fold (compared to SRM) by leveraging the large amount of data present in a SWATH-MS run
A new method to study protein complexes by chemical cross-linking
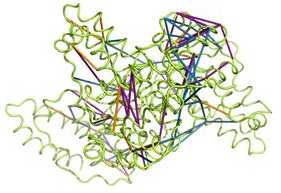
Researchers from the Aebersold group develop a complementary cross-linking chemistry that connects acidic residues in protein complexes.
Christen Lab, Microscopy, Asymmetric Protein Localization and Cell-Cycle Signalling in Bacteria
Investigate a novel protein localization factor
Christen Lab, Microscopy, Illuminating signaling networks in bacteria
Investigate how a secondary messenger triggers cellular heterogeneity
Christen Lab, Genomics, Quantitative Transposon Sequencing
Elucidate the core function of microbial genome by transposon insertion and NGS
Three new publications with contributions of the Sauer Lab
We congratulate the Hall lab (Biozentrum Basel) and the Lavigne/Briers group (KU Leuven)
Sauer Lab, Microbial metabolism, Metabolomics
A systems-level investigation of the stress-induced global metabolic adaptations in several organisms
Aebersold Lab, Proteomics, Cholesterol regulation
Analysis of cellular cholesterol regulation using targeted mass-spectrometry
Welcome to new post-doc
A warm welcome to Ilaria Piazza who joined our team as a post-doc.
Glyco-SWATH paper with Wlab participation published on MCP
Wlab contributed to a Glyco-SWATH project led by Dr. Yansheng Liu from Ruedi Aebersold's lab
Emanuela Milani selected to present her project at HUPO Congress in Madrid
The oral presentation on "AP-MS and BioID analysis of the HBx Viral-Host interactome" will be given at the Human Proteome Organisation (HUPO) 13th Annual World Congress in Madrid, Spain in the beginning of October. Congrats Ema!
Wlab contributed to a microbiome project led by Frank Schmidt
... from the University of Greifswald entitled "Comparative proteome analysis reveals conserved and specific adaptation patterns of Staphylococcus aureus after internalization by different types of human non-professional phagocytic host cells" which was just published in Frontiers in Microbiology. Congrats to Frank and his team!
Workshop on Computational Single Cell Biology at ISMB in Boston
We are organizing a workshop for the Special Interest Group on Computational Single Cell Biology that will take place in conjunction with ISMB in Boston, USA on July 12, 2014 (CSCB14).
Karel Novy selected for oral presentation at International Poxvirus, Asfarvirus & Iridovirus Conference
Karel Novy was selected for oral presentation of his abstract entitled "Potential Phosphoregulation of I7 Protease by H1 Phosphatase During Vaccinia Virus Morphogenesis" at the 2014 International Poxvirus, Asfarvirus & Iridovirus Conference at the end of September in Victoria, British Columbia, Canada. Congrats to Sam, Jason and Karel!
TRICEPS movie released!
Together with our commercial partner Dualsystems Biotech AG we had the opportunity to create a movie highlighting the patented TRICEPS-based LRC Technology which was developed by Andreas Frei during his PhD thesis in our lab.
Daniel Sévin wins Poster Award in Davos
Daniel Sévins wins 1 (out of 5) awards at the D-BIOL 2014 symposium in Davos.
LiP-SRM paper accepted in Nature Biotechnology
Congratulations to Yuehan, Paola et al. as their LiP-SRM paper was accepted in Nature Biotechnology
Poster prize for Andreas Kühne
Andreas Kühne won a Poster Prize at the D-BIOL Symposium in Davos. Congrats!
Welcome to new post-doc
A warm welcome to Simone Schopper who started a post-doc project in our lab.
A test for mass spectrometer fragmentation
The Aebersold lab developed a methodology for testing mass spectrometer fragmentation performance and improving data mining in targeted proteomics.
Cell size related changes in transcription and metabolism
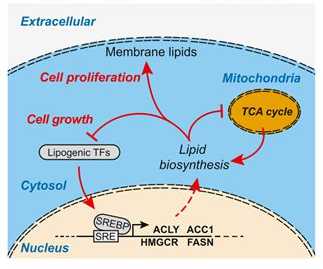
In collaboration with the Sauer lab, researcher from Dundee and Singapore University investigated mammalian cell size related differences in cytoskeletal and mitochondrial gene expression as well as in lipid and mitochondrial metabolism.
Bacterial metabolism during starvation
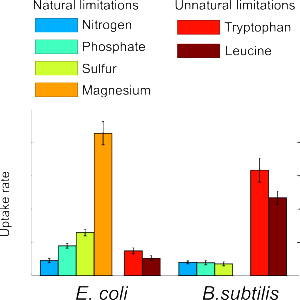
The Sauer Lab characterized metabolism under starvation conditions in two model bacteria, with one eye on ecology and one eye on biotechnology.
Novel metabolomics method for α-keto acids
Alpha-Keto acids (e.g. glyoxylate, oxaloacetate, pyruvate, ...) are highly reactive and thus unstable compounds. We established a new method for in situ stabilization and quantification by mass spectrometry. The technique also allows determination of labeling patterns.
Glucosamine supplementation extends life span
The Zamboni lab contributed to a study led by Prof. Ristow of ETH Zurich
Review - Principles of Metabolic Regulation
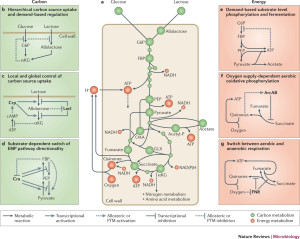
The Sauer Lab reviewed and delineated general principles of metabolic regulation by comparing the microbial model organisms E. coli, B. subtilis and S. cerevisiae.
Review - Kinetic models of metabolism
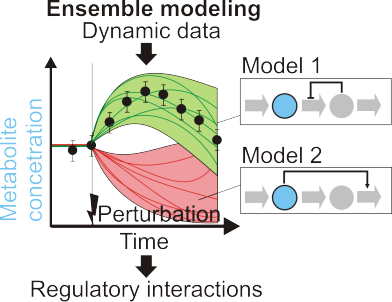
People from the Sauer group wrote a review about computational models of metabolism. The focus was on models that include reaction kinetics in order to simulate dynamics of metabolic networks. The finding is that such models experience a paradigm shift away from fitting parameters towards identifying key regulatory interactions.
DALCO cluster delivered to Wollscheid lab
DALCO computing and storage cluster arrived which takes our infrastructure for proteomic data processing to the next level. Awesome job in rigging the cluster for our specific needs Ulrich and Beat Rubischon!
Paper Published
Congrats to Juan and Paola: the paper Accumulation of oligomer-prone α-synuclein exacerbates synaptic and neuronal degeneration in vivo they contributed to was published in Brain!
New (PhD) students
A big welcome to our 'new' PhD student Pascal Leuenberger and semester students Marc van Oostrum and Carmen Schwarz!
Ubiquinone accumulation improves osmotic-stress tolerance in Escherichia coli
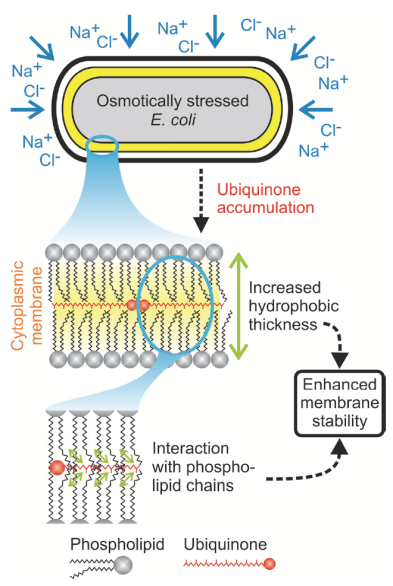
Using a novel metabolomics method, the Sauer group discovered a previously unknown mechanism bacteria use to cope with osmotic stress.
Dr. Leila Alexander starts as PostDoc
and will work on mechanisms of acquired drug resistance
Nontargeted Profiling of Coenzyme A thioesters
The Zamboni lab demonstrated and applied a method for selective and yet non-targeted profiling of Coenzyme A derivatives in cellular extracts
Paper Published
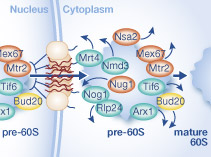
Congratulations to Andre and Paola. Their collaborative effort with the Panse group has resulted in a paper in Molecular Systems Biology: Targeted proteomics reveals compositional dynamics of 60S pre-ribosomes after nuclear export.
Daniel Sévin wins Poster Award at EMBO Symposium
Daniel received a Nature Reviews Microbiology Poster Award at the EMBO Symposium: New Approaches and Concepts in Microbiology in Heidelberg
Poster Award to Andreas Kühne
We congratulate Andreas Kühne for winning a Best Poster Award at the Conference on Systems Biology of Human Diseases in Heidelberg
Welcome Erasmus student
We welcome our Erasmus student Filip.
Review published

Martin Soste and Paola Picotti wrote a short review as part of a 'Highlights of Analytical Sciences in Switzerland' in CHIMIA about a complete mass-spectrometric map of a eukaryotic proteome.
Welcome semester students
We are welcoming our semester students Gea and Marta.
HUPO 2013
Wishing Andre, Martin and Yuehan good luck at HUPO 2013 and fun at the other side of the world!
The Christen lab starts its operations at IMSB
The newly appointed Assistant Professor Beat Christen joins IMSB
ERC grant for Paola
Many congratulations to Paola. She got awarded an ERC starting grant!
Paper accepted
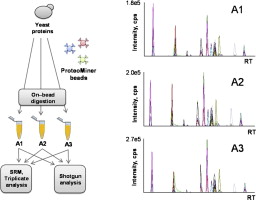
Congratulations to Francesco, Martin, Yuehan and Paola; their paper "Reproducibility of combinatorial peptide ligand libraries for proteome capture evaluated by selected reaction monitoring" was accepted for publication in Journal of Proteomics!
Targeted Proteomics named Method of the Year 2012
Nature Methods has realized the power and importance of targeted proteomics -in particular SRM based- and named it Method of the Year 2012.
Michael Zimmermann wins Poster awards at D-BIOL symposium
Congratulations!!
Mattia Zampieri wins Poster Award
at the Conference from Genomes to Systems in Heidelberg
Best Master Thesis
Daniel Sévin won a Prize for Best Master Thesis by the University of Mannheim for his work done in the Sauer Lab. Congratulations!
Jörg Büscher wins ETH Silver Medal
An ETH Silver medal was awarded to Jörg Büscher for his PhD Thesis under the supervision of Prof. U. Sauer
Karl Kochanowski wins Poster Prize
at the FEBS Advanced Course in Systems Biology in Innsbruck